Search Count: 48
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Ypel5
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: ZN |
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Nmnat1 Substrate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: NMN, ZN |
 |
Crystal Structure Of Human Fabp4 In Complex With 2-[1-(Methoxymethyl)Cyclopentyl]-6-Pentyl-4-Phenyl-3-(1H-Tetrazol-5-Yl)-5,6,7,8-Tetrahydroquinoline
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-03-06 Classification: LIPID BINDING PROTEIN Ligands: A1H4Y, DMS |
 |
N-Acetylglucosamine Kinase From Plesiomonas Shigelloides Compexed With Alpha-N-Acetylglucosamine-6-Phosphate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-08-10 Classification: SUGAR BINDING PROTEIN Ligands: EDO, PEG, 4QY, ZN, K, PO4, CL, TRS |
 |
N-Acetylglucosamine Kinase From Plesiomonas Shigelloides Compexed With Alpha-N-Acetylglucosamine And Amp-Pnp Inhibitor
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-08-10 Classification: SUGAR BINDING PROTEIN Ligands: ZN, ANP, PEG, PGE, NDG, EDO, K |
 |
N-Acetylglucosamine Kinase From Plesiomonas Shigelloides Compexed With Alpha-N-Acetylglucosamine
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-08-10 Classification: SUGAR BINDING PROTEIN Ligands: MPD, EDO, PGE, NDG, ZN, K, CL, PEG |
 |
Structure Of N-Acetylglucosamine Kinase From Plesiomonas Shigelloides In Complex With Amp-Pnp In The Absence Of N-Acetylglucoseamine Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-10 Classification: SUGAR BINDING PROTEIN Ligands: PEG, ANP, ZN, PGE, EDO |
 |
N-Acetylglucosamine Kinase From Plesiomonas Shigelloides Compexed With Alpha-N-Acetylglucosamine And Adp
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-08-03 Classification: SUGAR BINDING PROTEIN Ligands: ADP, GOL, IMD, NDG, ZN, EDO, PEG, K, IPA |
 |
Native Structure Of N-Acetylglucosamine Kinase From Plesiomonas Shigelloides
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Plesiomonas shigelloides 302-73
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-27 Classification: SUGAR BINDING PROTEIN Ligands: PEG, TRS, ZN, PGE, EDO, CL |
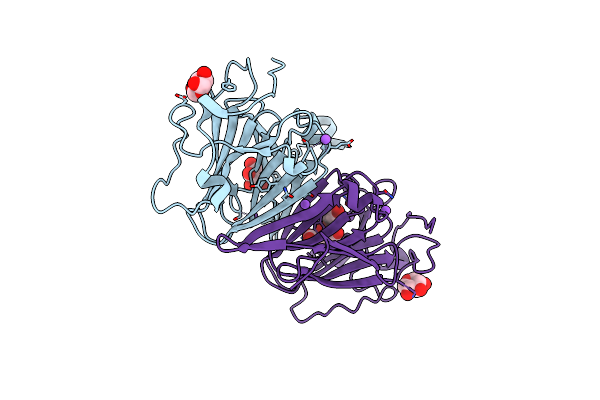 |
Organism: Coxiella burnetii (strain rsa 493 / nine mile phase i)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: XYP, XYS, NA, FLC |
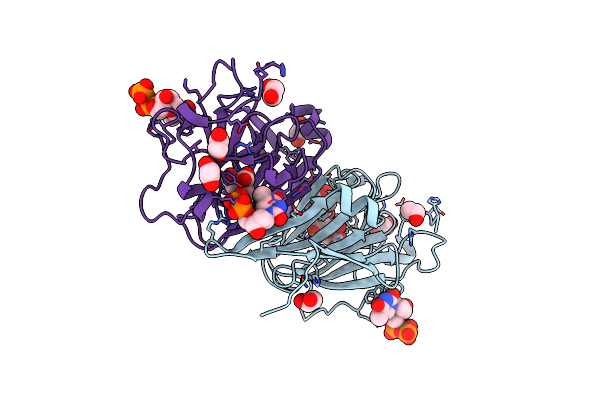 |
Organism: Coxiella burnetii (strain rsa 493 / nine mile phase i)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: TYD, CIT, EDO |
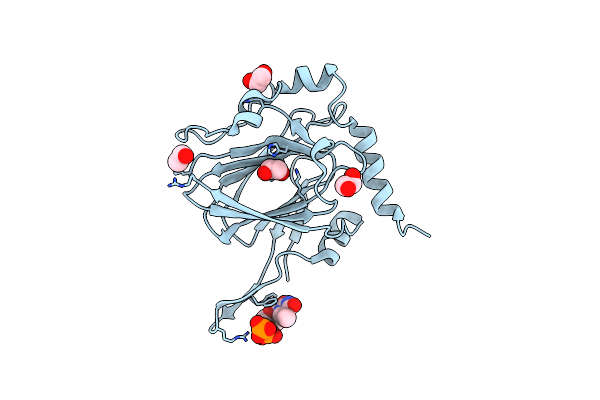 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: EDO, TYD, CL |
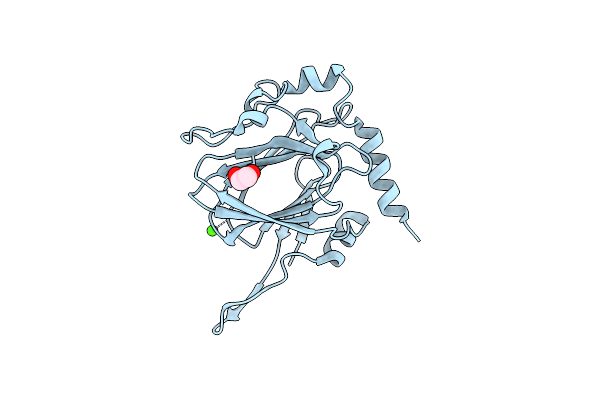 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: EDO, CA |
 |
Organism: Citrus unshiu, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2022-03-30 Classification: NUCLEAR PROTEIN Ligands: HOH |
 |
The Transcriptional Regulator Prfa From Listeria Monocytogenes In Complex With Inhibitor Of Wnt Production (Iwp)-2
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-27 Classification: TRANSCRIPTION Ligands: 9XK, DMS, NA |
 |
Structure Of Human Ketohexokinase-C In Complex With Fructose, No3, And Osthole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-07 Classification: TRANSFERASE Ligands: FRU, A0O, NO3 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: ADP, FRU, NO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-03-08 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: PG4, P6G, EDO, FMT, CA |