Search Count: 18
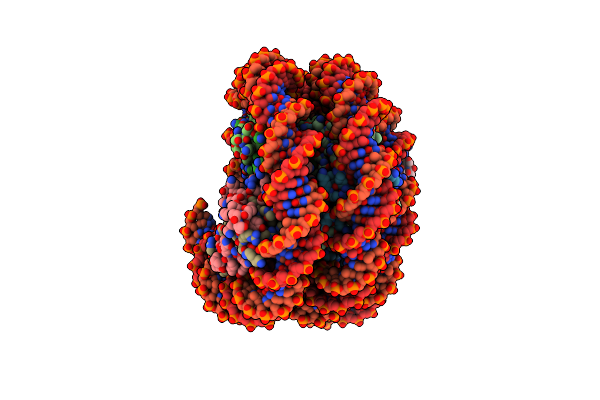 |
Structure Of A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN |
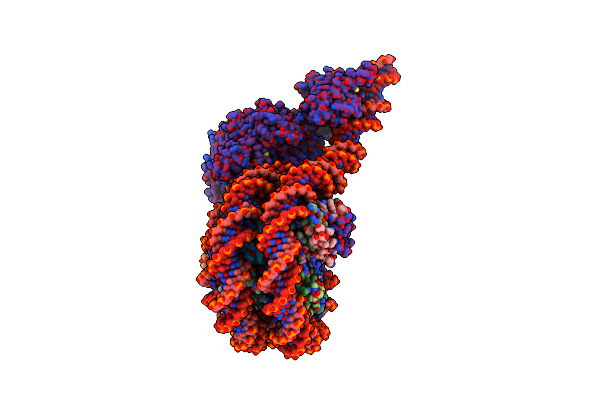 |
Structure Of Chd1 Bound To A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
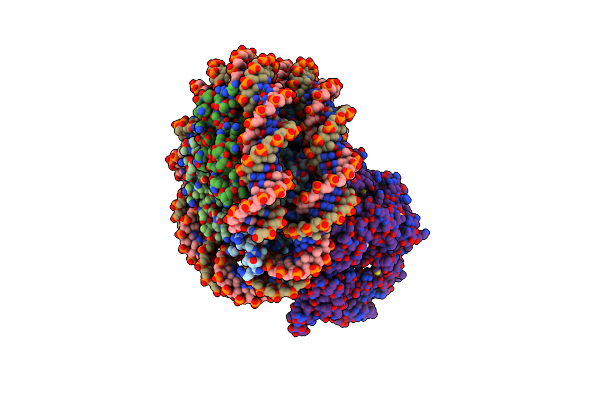 |
Structure Of Chd1 Bound To A Dinucleosome With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
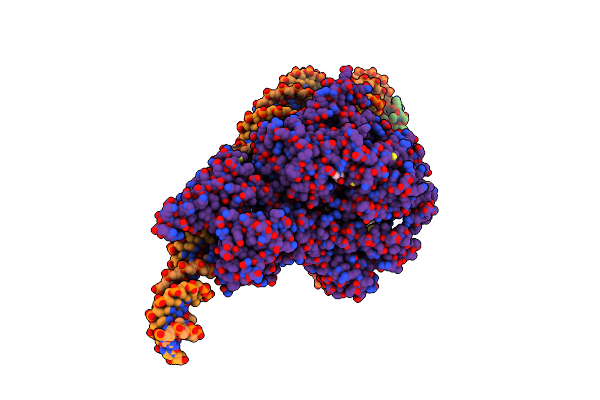 |
Structure Of A Mononucleosome Bound By One Copy Of Chd1 With The Dbd On The Exit-Side Dna.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4, DDS, MG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4, DDS |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4, DDS |
 |
Human Dna Polymerase Epsilon Bound To T-C Mismatched Dna (Post-Insertion State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4, DDS, CA |
 |
Human Dna Polymerase Epsilon Bound To T-C Mismatched Dna (Polymerase Arrest State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4 |
 |
Human Dna Polymerase Epsilon Bound To T-C Mismatched Dna (Frayed Substrate State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4 |
 |
Human Dna Polymerase Epsilon Bound To T-C Mismatched Dna (Mismatch Excision State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: REPLICATION Ligands: SF4, CA |
 |
Structure Of Human Mitochondrial Rnase P In Complex With Mitochondrial Pre-Trna-His(5,Ser)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN Ligands: NAD, ZN, SAH, MG |
 |
Structure Of Human Mitochondrial Rnase Z In Complex With Mitochondrial Pre-Trna-His(0,Ser)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN Ligands: NAD, ZN, SAH |
 |
Structure Of Human Mitochondrial Cca-Adding Enzyme In Complex With Mitochondrial Pre-Trna-Ile
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN Ligands: NAD, CDP, SAH |
 |
Structure Of Human Mitochondrial Mrpp1-Mrpp2 In Complex With Mitochondrial Pre-Trna-Ile
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN Ligands: NAD, SAH |
 |
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: ZN, CIT |
 |
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: ZN, CIT, ANP |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: ZN |

