Search Count: 13
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN Ligands: IMD, EDO |
 |
Organism: Sulfurivirga caldicuralii
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-09-07 Classification: LYASE |
 |
Organism: Rhodospirillaceae bacterium brh_c57
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-09-07 Classification: LYASE Ligands: CAP, MG |
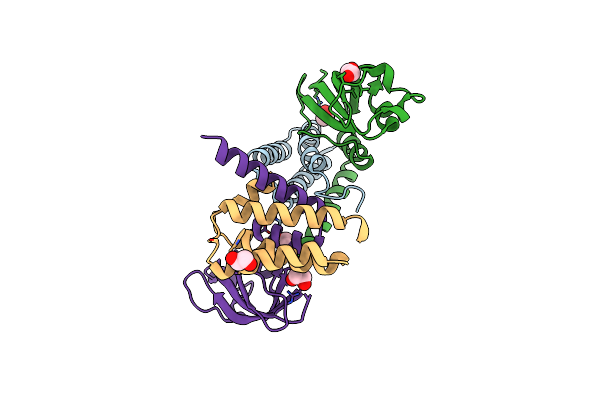 |
The 1.5 A Crystal Structure Of The Co-Factor Complex Of Nsp7 And The C-Terminal Domain Of Nsp8 From Sars Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: EDO |
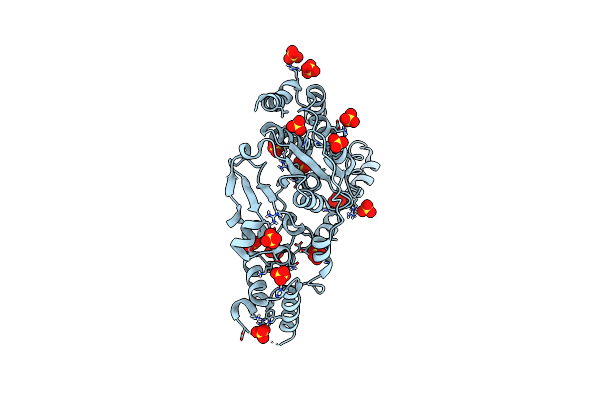 |
Organism: Homo sapiens, Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-17 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
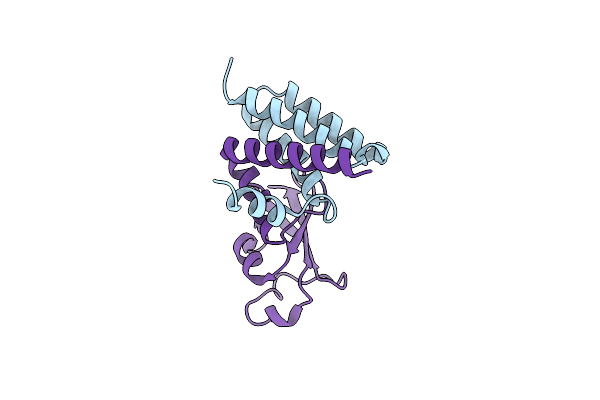
The 1.95 A Crystal Structure Of The Co-Factor Complex Of Nsp7 And The C-Terminal Domain Of Nsp8 From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-05-06 Classification: VIRAL PROTEIN Ligands: EDO |
 |
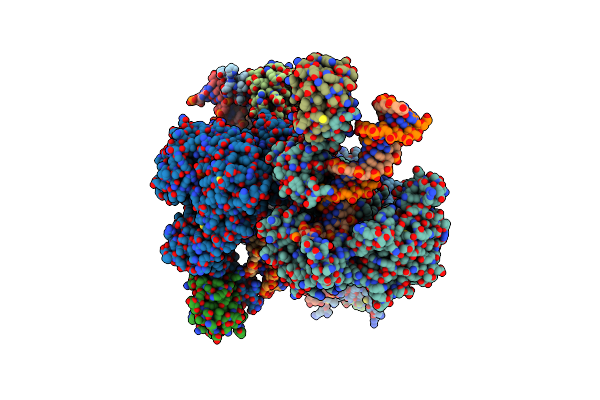
Crystal Structure Of The Co-Factor Complex Of Nsp7 And The C-Terminal Domain Of Nsp8 From Sars Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN |
 |
Structure Of T7 Dna Polymerase Bound To A Primer/Template Dna And A Peptide That Mimics The C-Terminal Tail Of The Primase-Helicase
Organism: Enterobacteria phage t7, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-03-04 Classification: TRANSFERASE/DNA BINDING PROTEIN/DNA Ligands: TTP, MG |
 |
Organism: Manduca sexta
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Manduca sexta
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Lachnospiraceae bacterium nd2006, Moraxella bovoculi
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Moraxella bovoculi, Lachnospiraceae bacterium nd2006
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |

