Search Count: 92
 |
Organism: Carica papaya
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: PO4, K |
 |
X-Ray Diffraction Structure Of Ctx-M-14 Beta-Lactamase Co-Crystallized With Avibactam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL, GOL, PO4 |
 |
X-Ray Diffraction Structure Of Ctx-M-14 Beta-Lactamase Soaked With Avibactam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL, PO4 |
 |
X-Ray Diffraction Structure Of The Ctx-M-14 Beta-Lactamase-Avibactam Complex An Inhibitor Cocktail-Soaked Crystal
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL, PO4 |
 |
Organism: Escherichia coli
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE |
 |
Microed Structure Of The Ctx-M-14 Beta-Lactamase-Avibactam Complex From Inhibitor Cocktail-Soaked Crystals
Organism: Escherichia coli
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL |
 |
Organism: Escherichia coli
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL |
 |
Microed Structure Of Ctx-M-14 Beta-Lactamase Co-Crystallized With Avibactam
Organism: Escherichia coli
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NXL |
 |
X-Ray Diffraction Structure Of Lysozyme Co-Crystallized With N,N',N"-Triacetylchitotriose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: CL, NA |
 |
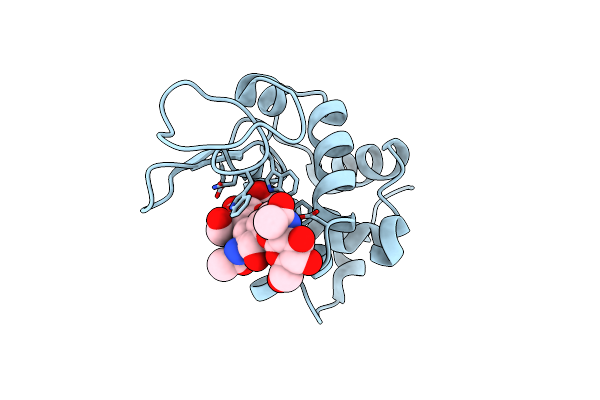
X-Ray Diffraction Structure Of Lysozyme Complexed With N,N',N"-Triacetylchitotriose From A Cocktail-Soaked Crystal
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: CL |
 |
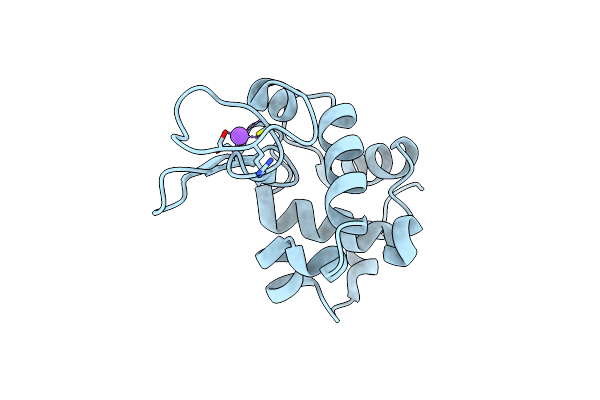
X-Ray Diffraction Structure Of Lysozyme Soaked With N,N',N"-Triacetylchitotriose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: CL |
 |
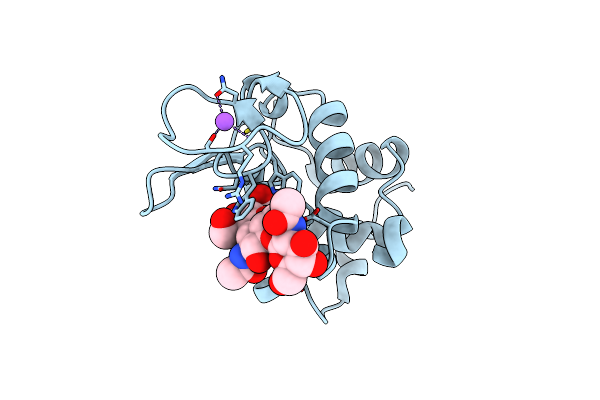
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE |
 |
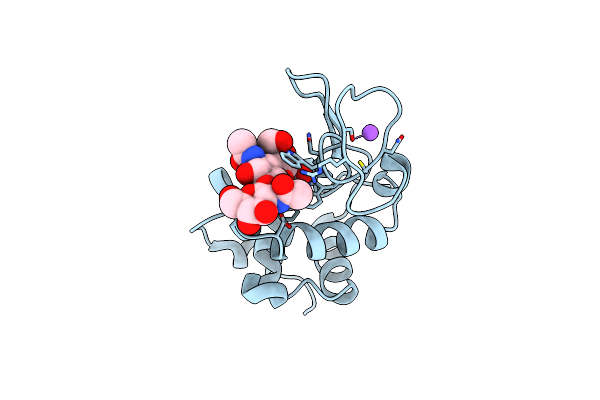
Microed Structure Of Lysozyme Complexed With N,N',N"-Triacetylchitotriose From Cocktail-Soaked Crystals
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE |
 |
Microed Structure Of Lysozyme Co-Crystallized With N,N',N"-Triacetylchitotriose
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: HOH |
 |
Structural Asymmetry In Sars-Cov-2 Nsp15 Hexamer Important For Catalytic Activity
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN |

