Search Count: 59
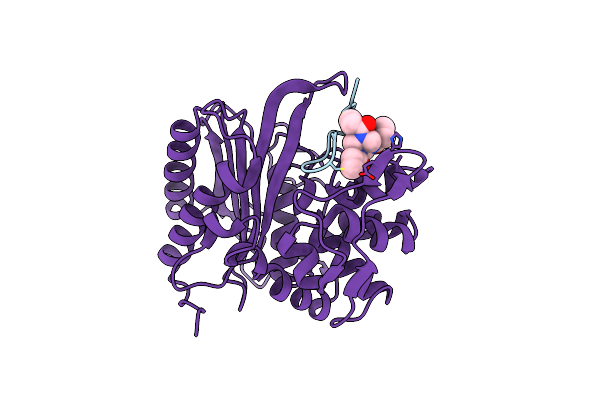 |
The Structure Of E. Coli Penicillin Binding Protein 3 (Pbp3) In Complex With A Bicyclic Peptide Inhibitor
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-04-03 Classification: HYDROLASE Ligands: 29N |
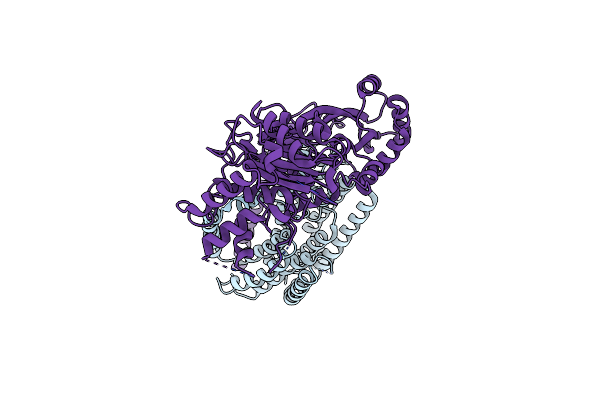 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Citrobacter rodentium
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: ZN, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: ZN |
 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Ligand Bound State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: GPP |
 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Apo State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
Structure-Function Analyses Of Dual-Bon Domain Protein Dolp Identifies Phospholipid Binding As A New Mechanism For Protein Localisation To The Cell Division Site
Organism: Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2020-12-30 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-04 Classification: PROTEIN BINDING |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-09-09 Classification: ANTIMICROBIAL PROTEIN |
 |
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, PO4, EDO |
 |
Crystal Structure Of S. Aureus Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, EDO, PEG, MG |
 |
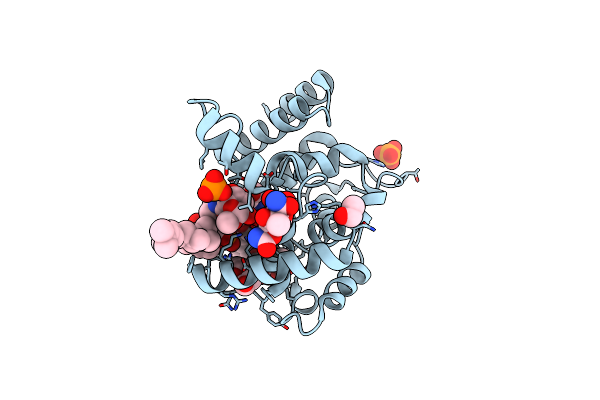
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To A Seryl Sulfamoyl Adenosine Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: JPE, CL, SO4, DMS, EDO, TRS |
 |
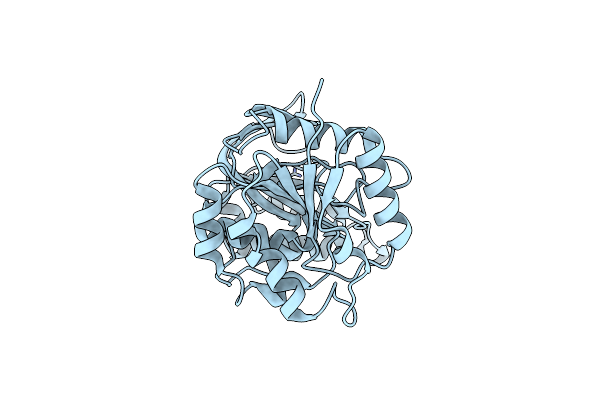
Staphylococcus Aureus Monofunctional Glycosyltransferase In Complex With Moenomycin
Organism: Staphylococcus aureus mw2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-06-27 Classification: TRANSFERASE Ligands: M0E, EDO, PO4, 1QW |
 |
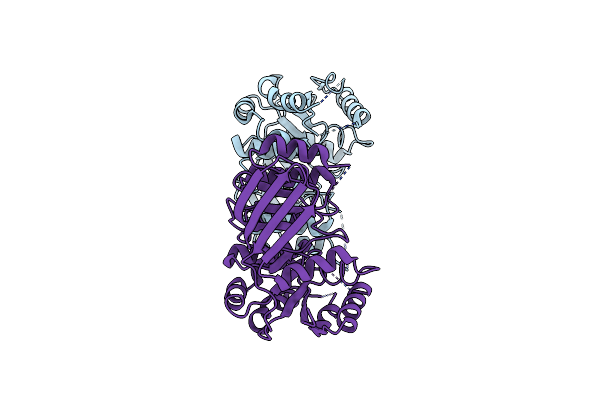
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: MG |
 |
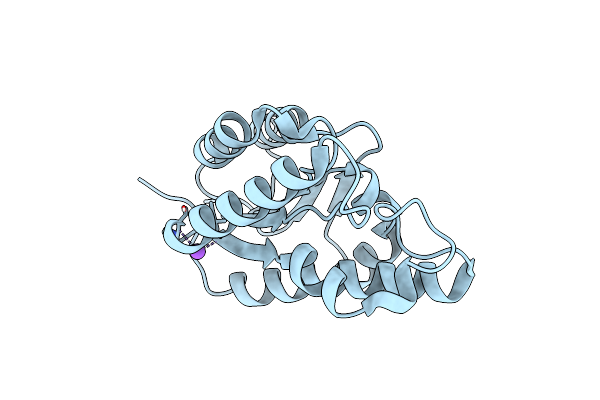
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2018-04-25 Classification: HYDROLASE |
 |
Structure Of Streptococcus Pneumoniae Peptidoglycan O-Acetyltransferase A (Oata) C-Terminal Catalytic Domain
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2017-10-25 Classification: TRANSFERASE |
 |
Structure Of Streptococcus Pneumoniae Peptidoglycan O-Acetyltransferase A (Oata) C-Terminal Catalytic Domain With Methylsulfonyl Adduct
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-25 Classification: TRANSFERASE Ligands: 03S, NA |
 |
Crystal Structure Of The Autolysin Lyta From Streptococcus Pneumoniae Tigr4
Organism: Streptococcus pneumoniae tigr4
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-27 Classification: HYDROLASE Ligands: GOL, ZN, CHT |
 |
The X-Ray Crystal Structures Of D-Alanyl-D-Alanine Ligase In Complex Adp And D-Cycloserine Phosphate
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-01-21 Classification: LIGASE Ligands: ADP, DS0, MG, GOL |

