Search Count: 18
 |
Organism: Gut metagenome
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Gut metagenome
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: PO4, BGC |
 |
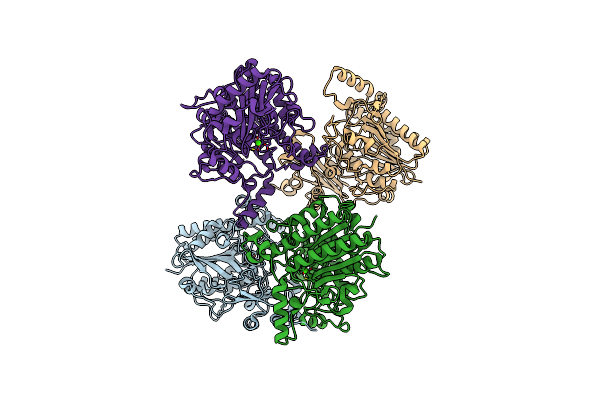
The Cryo-Em Structure Of Bacterial Beta-1,3-Glucan Phosphorylase From Family Gh161
Organism: Gut metagenome
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
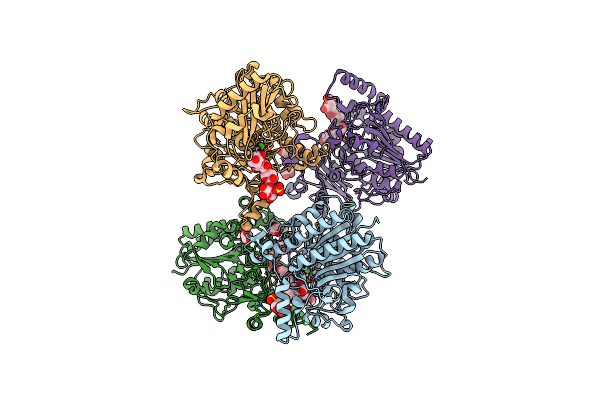
Crystal Structure Of The Family S1_19 Carrageenan Sulfatase Zgcgsa From Zobellia Galactanivorans In Complex With Hybrid A-I-Neocarratetraose
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: CA, BR, CL, GOL |
 |
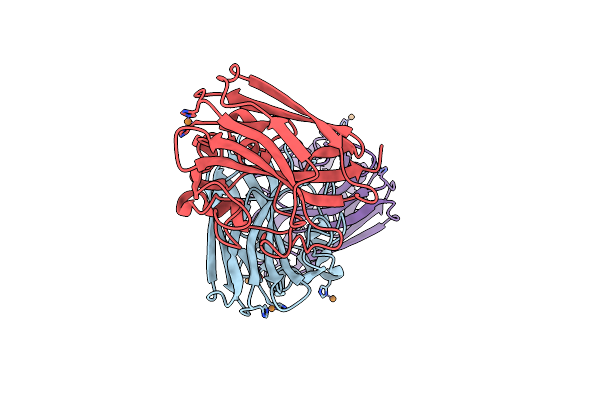
Structure Of The Apo Zgcgsa Carrageenan-Sulfatase (S1_19) From The Marine Bacterium Zobellia Galactanivorans
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: BR, CL, CA |
 |
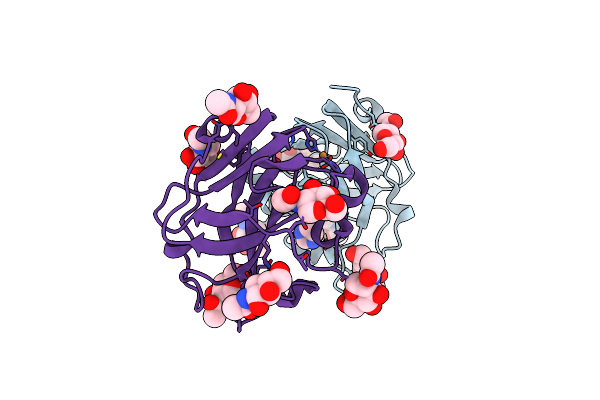
Crystal Structure Of The Family S1_19 Carrageenan Sulfatase Zgcgsa From Zobellia Galactanivorans In Complex With Hybrid B-K-Neocarratetraose
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: CA, BR, CL, GOL |
 |
Organism: Photorhabdus laumondii subsp. laumondii (strain dsm 15139 / cip 105565 / tt01)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Laetisaria arvalis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-11-13 Classification: METAL BINDING PROTEIN Ligands: NAG, CU, IMD |
 |
Organism: Laetisaria arvalis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-11-13 Classification: METAL BINDING PROTEIN Ligands: CU, NAG, 1PE, PGE, IMD |
 |
Organism: Laetisaria arvalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-11-13 Classification: METAL BINDING PROTEIN Ligands: NAG, CU |
 |
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: MG, GOL, EDO |
 |
Crystal Structure Of A Xylan-Active Lytic Polysaccharide Monooxygenase From Pycnoporus Coccineus.
Organism: Pycnoporus cinnabarinus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2018-01-17 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of The 3,6-Anhydro-D-Galactonate Cycloisomerase From Zobellia Galactanivorans
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-12-06 Classification: ISOMERASE Ligands: MG |
 |
A 3,6-Anhydro-D-Galactosidase Produced By Zobellia Galactanivorans. This Is An Exo-Lytic Enzyme That Hydrolyzes Terminal 3,6-Anhydro-D-Galactose From The Non-Reducing End Of Carrageenan Oligosaccharides.
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-11-29 Classification: HYDROLASE Ligands: MPD, TRS, CL |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-03-06 Classification: HYDROLASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-02-13 Classification: HYDROLASE Ligands: GOL, CL |
 |
Organism: Ruminococcus gnavus e1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-09-28 Classification: HYDROLASE Ligands: GOL, EDO, PO4, MG |
 |
Galactosidase Domain Of Alpha-Galactosidase-Sucrose Kinase, Agask, In Complex With Galactose
Organism: Ruminococcus gnavus e1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2011-09-28 Classification: HYDROLASE Ligands: GOL, PGE, PO4, NA, GLA, GAL |

