Search Count: 29
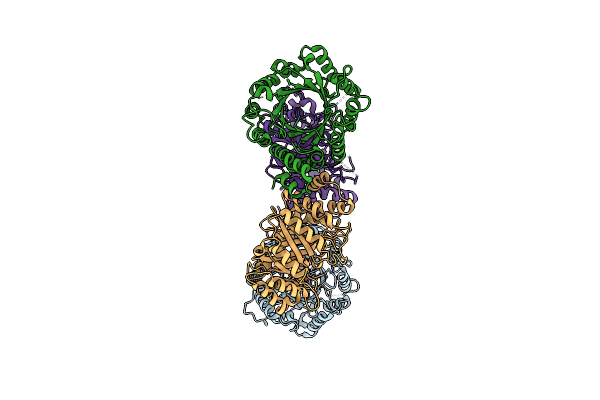 |
Crystal Structure Of Dmats1 Prenyltransferase In Complex With L-Tyr And Dmspp
Organism: Fusarium fujikuroi
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: TYR, DST |
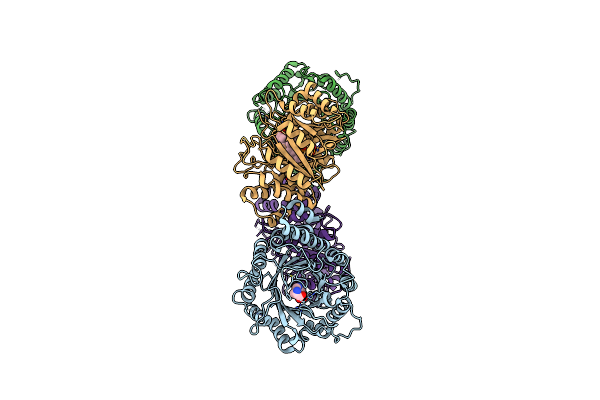 |
Crystal Structure Of Dmats1 Prenyltransferase In Complex With L-Trp And Gspp
Organism: Fusarium fujikuroi
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: TRP, GST |
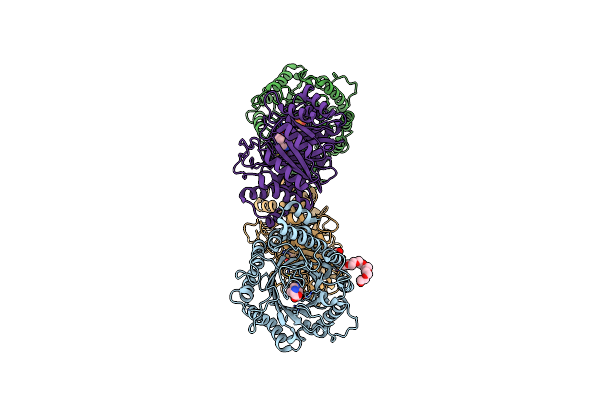 |
Crystal Structure Of Dmats1 Prenyltransferase In Complex With L-Trp And Dmspp
Organism: Fusarium fujikuroi
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: TRP, PE8, DST, EDO |
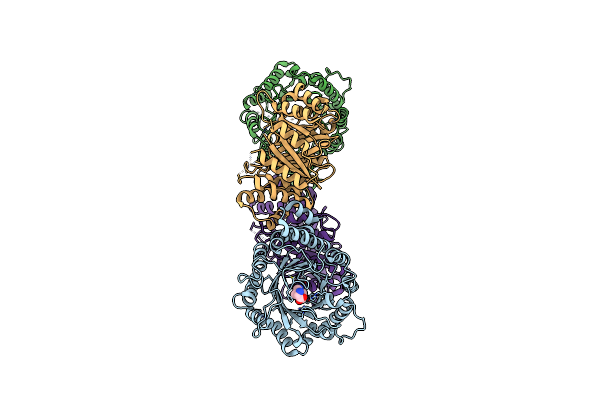 |
Organism: Fusarium fujikuroi
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: TRP |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 5-[(3-Aminopropyl)Amino]Pentane-1-Thiol
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: MES, SS9, ZN, MG, K |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 5-[(3-Aminopropyl)Amino]Pentylboronic Acid
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: OKP, K, MG, ZN |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 7-[(3-Aminopropyl)Amino]-1,1,1-Trifluoroheptan-2-One
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: K, ZN, MG, FKS |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 4-(Dimethylamino)-N-[7-Hydroxyamino)-7-Oxoheptyl]Benzamide
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: ZN, K, MG, B3N |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With Acetate
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: ACT, EDO, ZN, MG, K |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 6-[(3-Aminopropyl)Amino]-N-Hydroxyhexanamide
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: ZN, K, MG, EDO, XS6 |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 6-Amino-N-Hydroxyhexanamide
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: ZN, K, MG, 6XA |
 |
Crystal Structure Of Marinobacter Subterrani Acetylpolyamine Amidohydrolase (Msapah) Complexed With 8-Amino-N-Hydroxyoctanamide
Organism: Marinobacter subterrani
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-09-18 Classification: HYDROLASE/INHIBITOR Ligands: ZN, MG, K, OKS |
 |
Organism: Salinispora arenicola (strain cns-205)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-07-17 Classification: TRANSFERASE Ligands: B3P, TRS |
 |
Organism: Salinispora arenicola (strain cns-205)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2019-07-17 Classification: TRANSFERASE Ligands: TRP, BEZ, DPO, IMD |
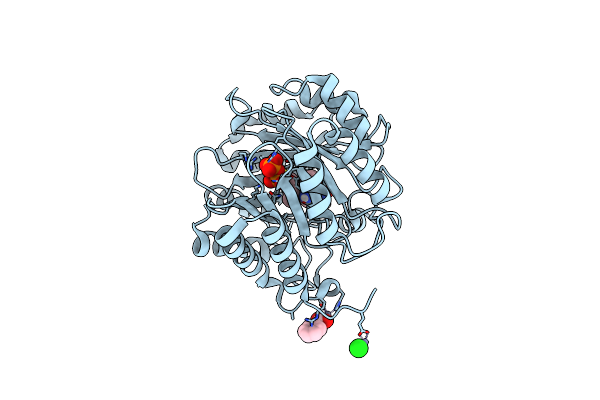 |
Crystal Structure Of Cymd Prenyltransferase Complexed With L-Tryptophan And Dmspp
Organism: Salinispora arenicola (strain cns-205)
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-07-17 Classification: TRANSFERASE Ligands: DST, TRP, ZN, CL, BEZ |
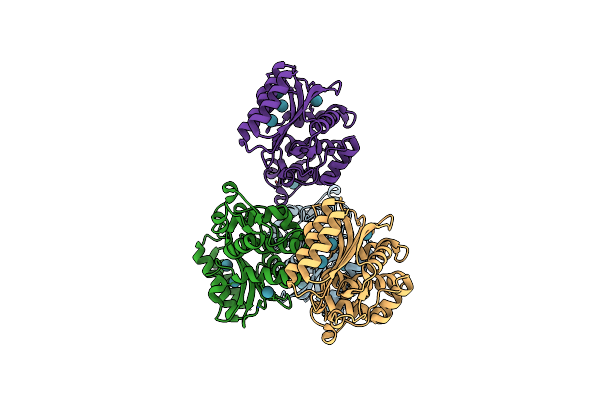 |
Crystal Structure Of Tem1 Beta-Lactamase Mutant I263A In The Presence Of 1.2 Mpa Xenon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: XE |
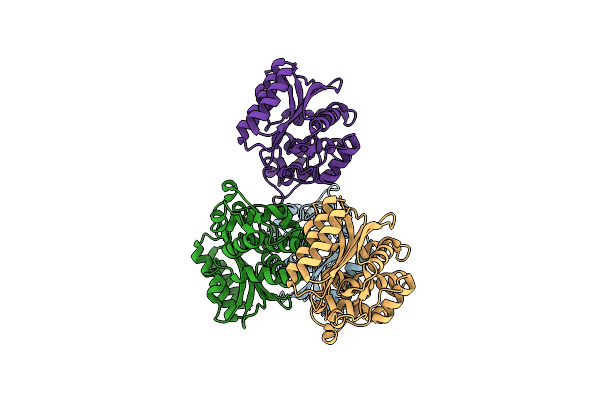 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-08-22 Classification: HYDROLASE |
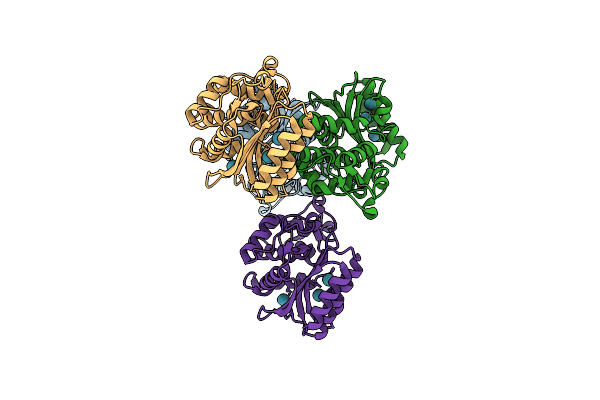 |
Crystal Structure Of Tem1 Beta-Lactamase Mutant I263L In The Presence Of 1.2 Mpa Xenon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: XE |
 |
Crystal Structure Of Ferritin E65R Mutant From Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2017-06-28 Classification: HYDROLASE |

