Search Count: 24
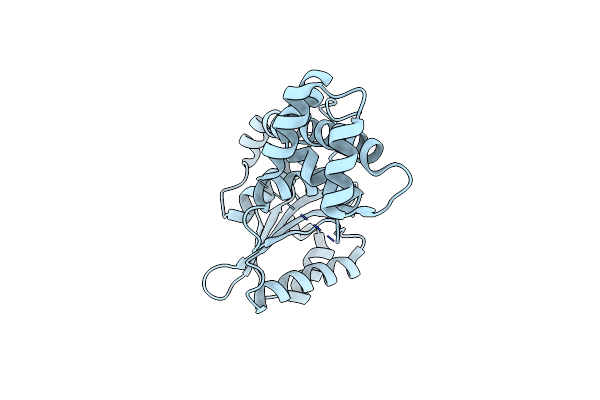 |
Crystal Structure Of The Deubiquitylase Domain From The Orientia Tsutsugamushi Protein Ott_1962 (Otdub)
Organism: Orientia tsutsugamushi (strain ikeda)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-01 Classification: HYDROLASE |
 |
Crystal Structure Of The Orientia Tsutsugamushi Otdub In Complex With Three Molecules Of Ubiquitin
Organism: Orientia tsutsugamushi (strain ikeda), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-01 Classification: HYDROLASE |
 |
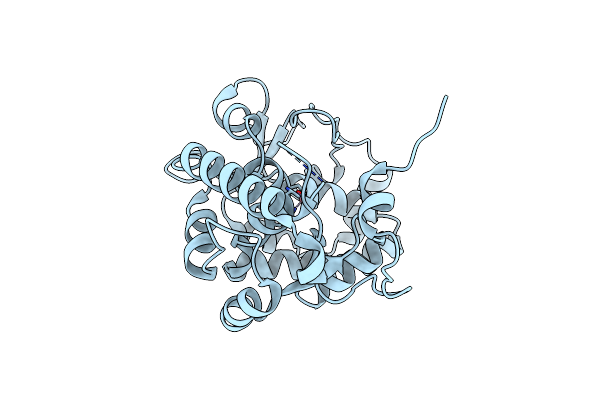
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-02-18 Classification: OXIDOREDUCTASE Ligands: CO, EDO |
 |
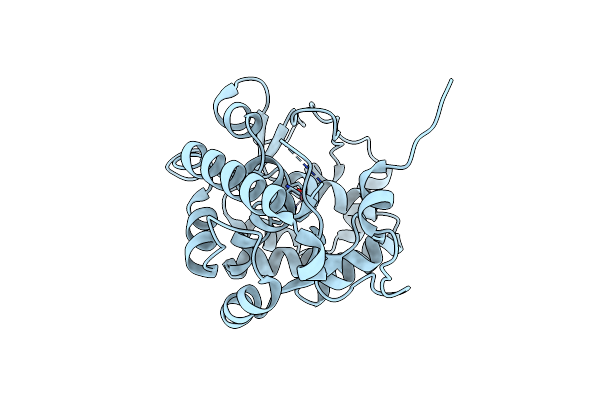
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-02-18 Classification: OXIDOREDUCTASE Ligands: CO |
 |
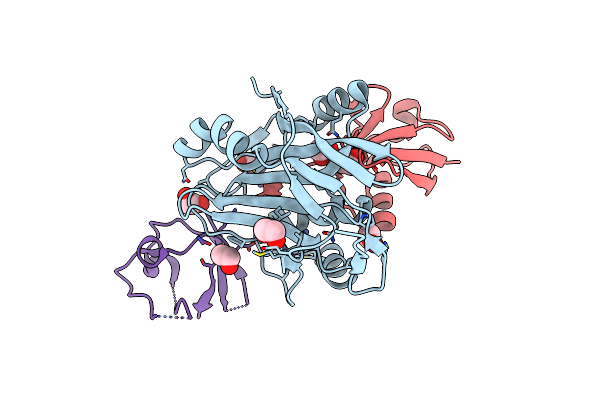
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-02-18 Classification: OXIDOREDUCTASE Ligands: CO |
 |
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-02-18 Classification: OXIDOREDUCTASE Ligands: CO |
 |
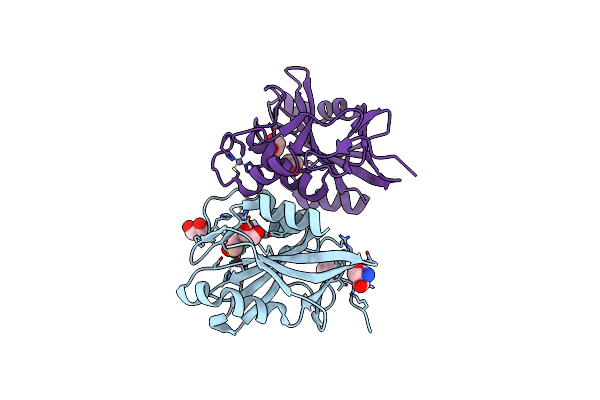
The Crystal Structure Of The Dub Domain Of Amsh Orthologue, Sst2 From S. Pombe, In Complex With Lysine 63-Linked Diubiquitin
Organism: Schizosaccharomyces pombe, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-10-08 Classification: HYDROLASE/PROTEIN BINDING Ligands: ZN, EDO |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-06-18 Classification: HYDROLASE Ligands: ZN, EDO, TRS |
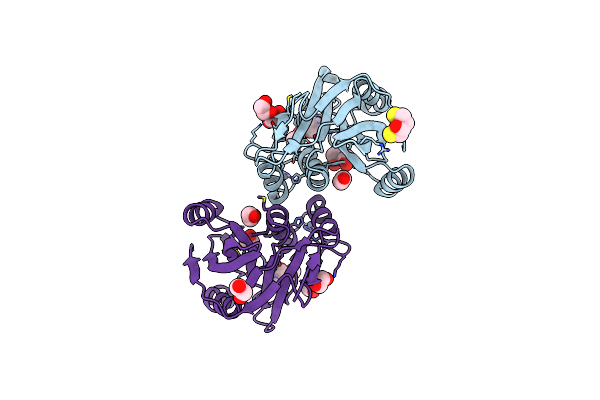 |
Crystal Structure Of Schizosaccharomyces Pombe Amsh-Like Protein Sst2 T319I Mutant
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-06-18 Classification: HYDROLASE Ligands: ZN, EDO, DTT |
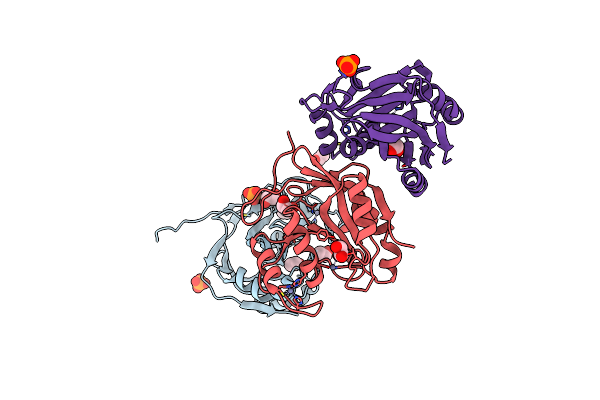 |
Crystal Structure Of S. Pombe Amsh-Like Protease Sst2 Catalytic Domain From P212121 Space Group
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-18 Classification: HYDROLASE Ligands: EDO, PO4, GLY, ZN |
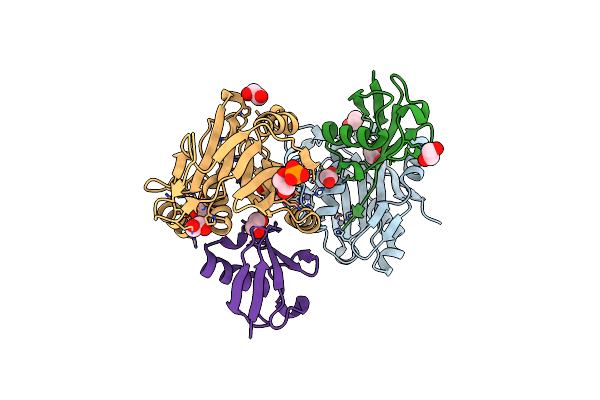 |
Crystal Structure Of Schizosaccharomyces Pombe Amsh-Like Protease Sst2 E286A Mutant Bound To Ubiquitin
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2014-06-18 Classification: HYDROLASE/PROTEIN BINDING Ligands: ZN, EDO, PO4 |
 |
Crystal Structure Of Schizosaccharomyces Pombe Amsh-Like Protease Sst2 Catalytic Domain Bound To Ubiquitin
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-06-18 Classification: HYDROLASE/PROTEIN BINDING Ligands: ZN, EDO, PO4 |
 |
Insights Into The Mechanism Of Deubiquitination By Jamm Deubiquitinases From Co-Crystal Structures Of Enzyme With Substrate And Product
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-06-18 Classification: Hydrolase/transcription Ligands: ZN, EDO |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-04-30 Classification: HYDROLASE Ligands: ZN, EDO, TRS |
 |
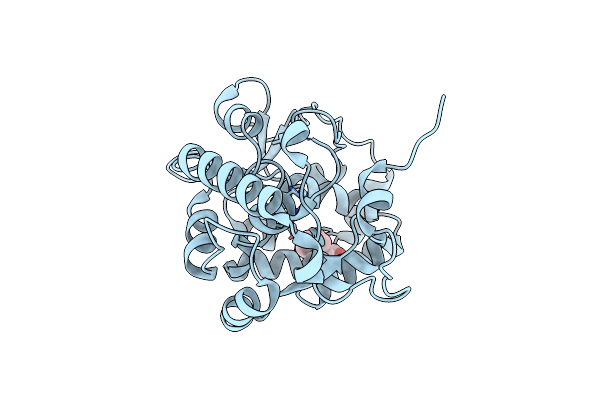
Crystal Structure Of Schizosaccharomyces Pombe Sst2 Catalytic Domain And Ubiquitin
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: ZN, CL, EDO |
 |
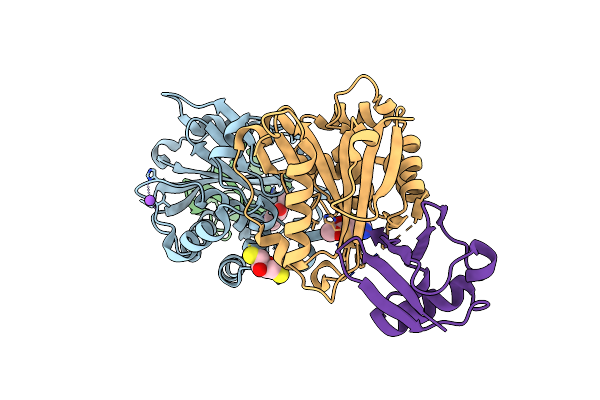
Crystal Structure Of Phenylalanine Hydroxylase S203P Mutant From Chromobacterium Violaceum
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: PHE, CO |
 |
Iron And Phenylalanine Bound Crystal Structure Of Phenylalanine Hydroxylase From Chromobacterium Violaceum
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: FE, PHE |
 |
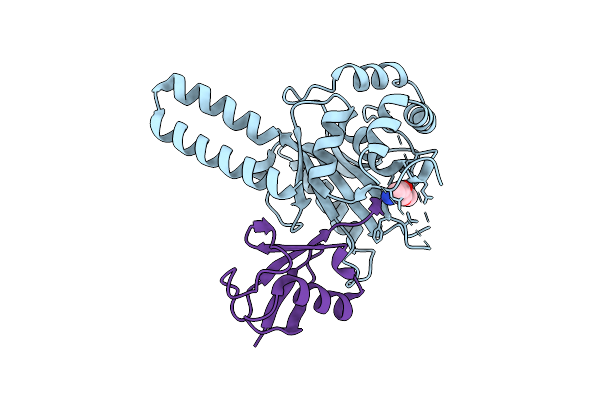
Crystal Structure Of Trichinella Spiralis Uch37 Catalytic Domain Bound To Ubiquitin Vinyl Methyl Ester
Organism: Trichinella spiralis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-05-29 Classification: Hydrolase/Signaling Protein Ligands: DTT, NA, GVE |
 |
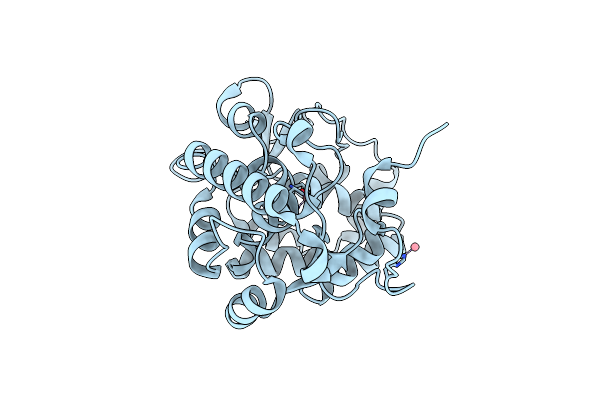
Crystal Structure Of Trichinella Spiralis Uch37 Bound To Ubiquitin Vinyl Methyl Ester
Organism: Trichinella spiralis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-29 Classification: Hydrolase/Signaling Protein Ligands: GVE |
 |
Crystallographic Structure Of Phenylalanine Hydroxylase From Chromobacterium Violaceum Y155A Mutation
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: CO |

