Search Count: 20
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: Replication/DNA/RNA Ligands: ZN, MG, DCP |
 |
Organism: Acetivibrio thermocellus dsm 1313
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-05-15 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: EDO |
 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester, Sp2-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, PO4, EDO, PGE |
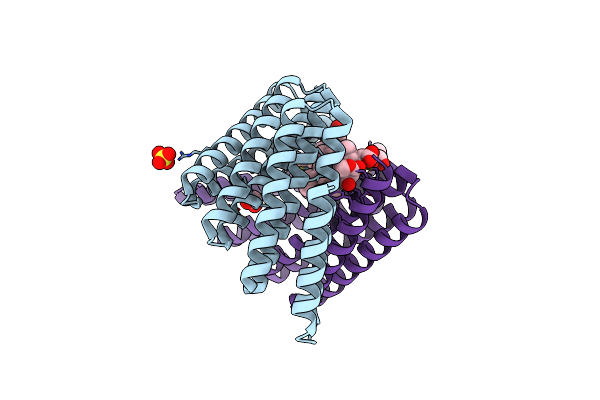 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester Matching Geometry Of Purple Bacterial Special Pair, Sp1-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, SO4, EDO |
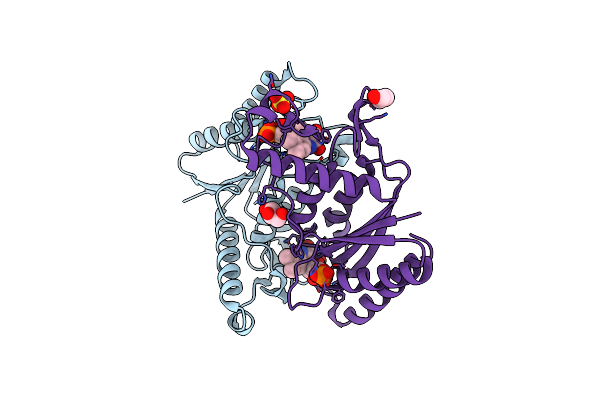 |
Organism: Bacillus okhensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, SO4 |
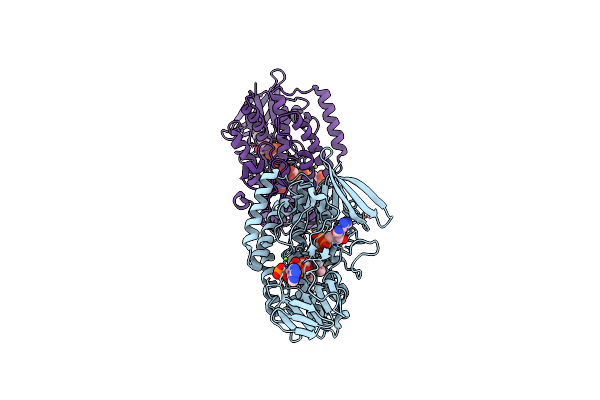 |
Structure Of Nitrincola Lacisaponensis Flavin-Containing Monooxygenase (Fmo) In Complex With Fad And Nadp+
Organism: Nitrincola lacisaponensis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-01-02 Classification: FLAVOPROTEIN Ligands: FAD, NAP, MG |
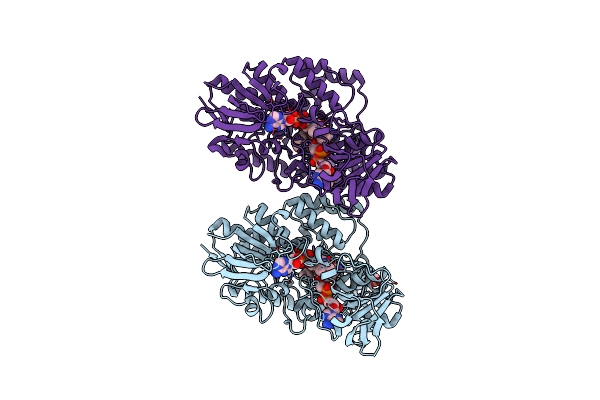 |
Thermocrispum Municipale Cyclohexanone Monooxygenase Bound To Hexanoic Acid
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-12 Classification: FLAVOPROTEIN Ligands: FAD, NAP, 6NA, ACT |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Crystal Structure Of The V465T Mutant Of 5-(Hydroxymethyl)Furfural Oxidase (Hmfo)
Organism: Methylovorus sp. (strain mp688)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-07-26 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Crystal Structure Of Trans-Membrane Domain Of Cylindrospermum Stagnale Nadph-Oxidase 5 (Nox5)
Organism: Cylindrospermum stagnale pcc 7417
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-06-28 Classification: MEMBRANE PROTEIN Ligands: SO4, HEM, LFA, D10, GOL, PEG |
 |
Crystal Structure Of Dehydrogenase Domain Of Cylindrospermum Stagnale Nadph-Oxidase 5 (Nox5)
Organism: Cylindrospermum stagnale pcc 7417
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-06-28 Classification: OXIDOREDUCTASE Ligands: FAD, CL, P6G, PEG, GOL |
 |
Cyclohexanone Monooxygenase From T. Municipale: Reduced Enzyme Bound To Nadp+
Organism: Thermocrispum municipale dsm 44069
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: FAD, NA7, GOL |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From Thermocrispum Municipale In The Oxidised State With A Bound Nicotinamide.
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, NCA, TRS, GOL |
 |
Organism: Streptococcus sp. 'group a'
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-05-25 Classification: OXIDOREDUCTASE |

