Search Count: 251
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: TRANSPORT PROTEIN Ligands: 377, GLU, CYZ |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: GLU, CYZ |
 |
Crystal Structure Of A Serine Protease Inhibitor Hpi From Hevea Brasiliensis
Organism: Hevea brasiliensis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-30 Classification: PLANT PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GOL, ACT, PEG |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GLA, MG |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: A2G, GOL, A1AAJ, ACT, PEG |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: CA, ACT, GOL |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GLA, EPE, PG4, CA, ACT, GOL |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA |
 |
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GOL, ACT, PEG, NI |
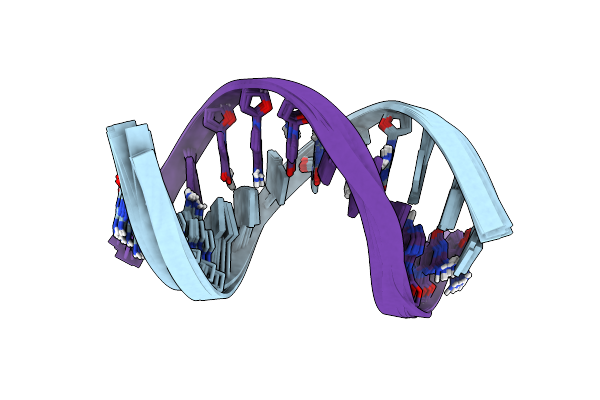 |
Solution Structure Of A Silver Ion Mediated Dna Duplex With Universal 7-Deazapurine Substitutions
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2024-09-18 Classification: DNA Ligands: AG |
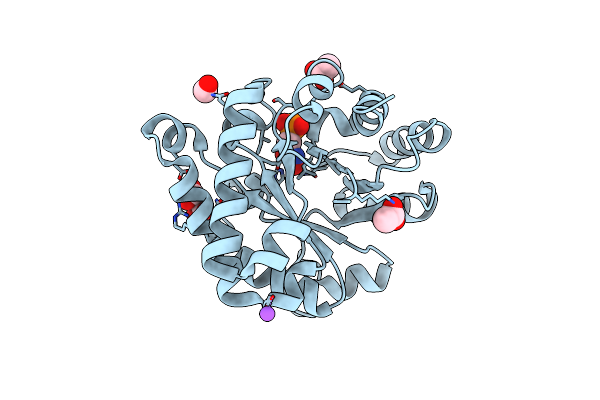 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-09-04 Classification: ISOMERASE |
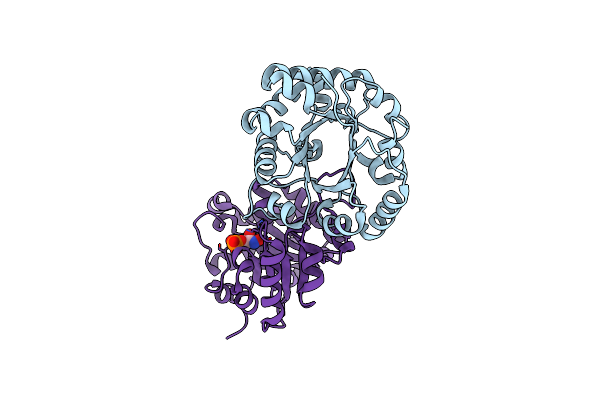 |
Crystal Structure Of The Worst Case Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH |
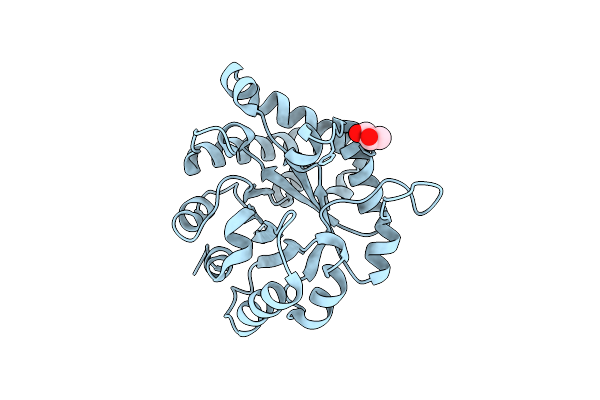 |
Crystal Structure Of The Ancestral Triosephosphate Isomerase Reconstruction Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: GOL |
 |
Crystal Structure Of The Reconstruction Of The Worst Case Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH, ACY, PGE, ACT, NA, EDO, PEG, CL |
 |
Crystal Structure Of The Worst Case Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: F |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-08-14 Classification: HYDROLASE |

