Search Count: 15
 |
Crystal Structure Of Arsenite Oxidase From Pseudorhizobium Banfieldiae Str. Nt-26 (Nt-26 Aio) Bound To Antimony Trioxide
Organism: Pseudorhizobium banfieldiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: MGD, O, 4MO, F3S, SO4, PGE, GOL, SBO, P33, FES |
 |
Crystal Structure Of Arsenite Oxidase From Alcaligenes Faecalis (Af Aio) Bound To Arsenite
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: MGD, MO, F3S, PGE, GOL, AST, EDO, PEG, IPA, FES |
 |
Crystal Structure Of Arsenite Oxidase From Alcaligenes Faecalis (Af Aio) Bound To Antimony Oxyanion
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: MGD, O, 4MO, F3S, PEG, ACT, PO4, UJI, CL, GOL, TRS, FES, EDO, SO4, NA |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Methione Oxide Ligand (Complex 6B)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, RU, CMO, GOL |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Pyridine Ligand (Complex 7)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, FMT, CMO, RU |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Pyridine Ligand (Complex 8)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, RU, CMO |
 |
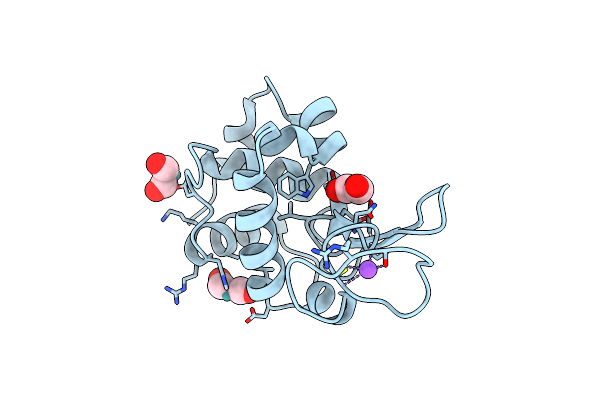
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-02-06 Classification: HYDROLASE Ligands: K3G, NA, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2011-01-26 Classification: HYDROLASE Ligands: RU, NA, CMO, CL |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
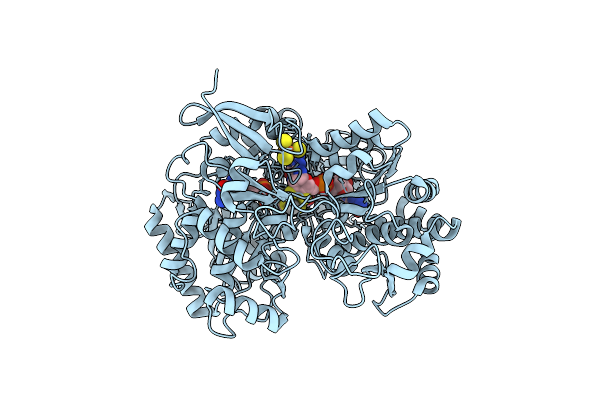
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX |
 |
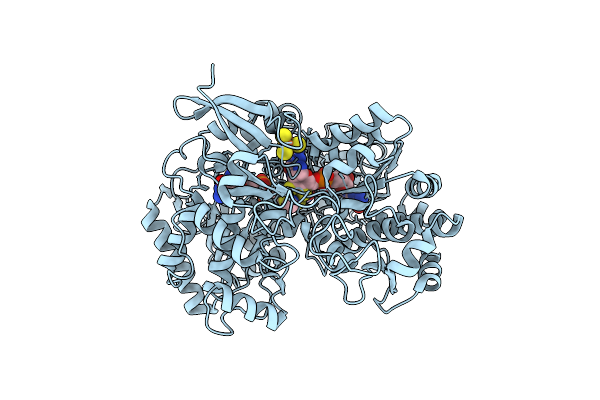
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, NO2, NO3 |
 |
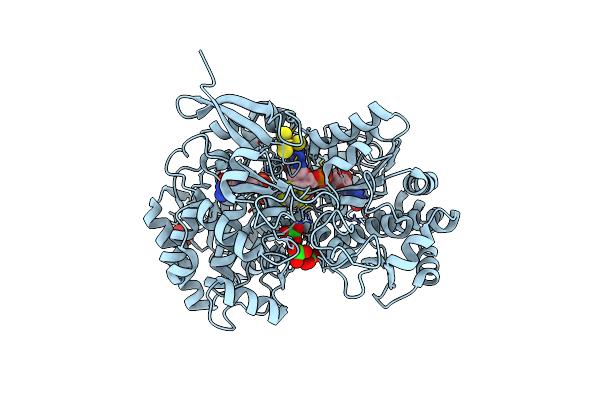
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, CYN, NO2 |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, LCP |
 |
A New Catalytic Mechanism Of Periplasmic Nitrate Reductase From Desulfovibrio Desulfuricans Atcc 27774 From Crystallographic And Epr Data And Based On Detailed Analysis Of The Sixth Ligand
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-03-18 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, UNX, LCP |

