Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-5-Methyl-Trp In Complex With Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: ZN, A1BNI, CL, NA |
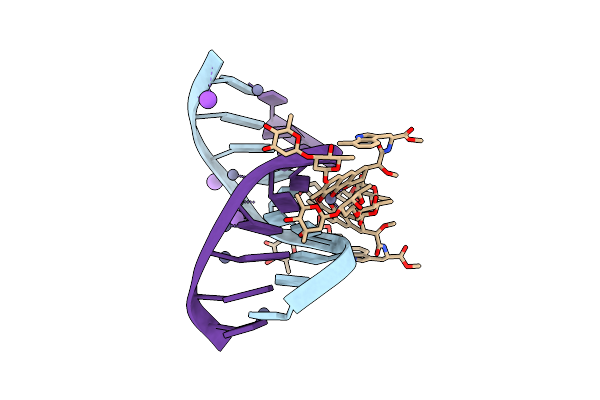 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-7-Methyl-Trp In Complex With Double-Stranded Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: A1BNK, ZN, NA |
 |
Crystal Structure Of The Dna Binding Domain Of Human Transcription Factor Fli1 In Complex With 16-Mer Dna Cagaggatgtggcttc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2020-11-25 Classification: TRANSCRIPTION |
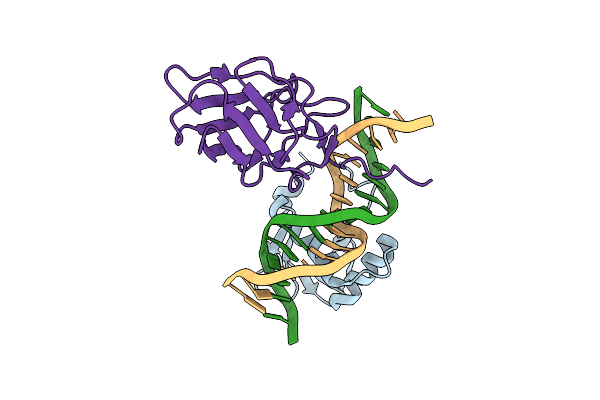 |
Crystal Structure Of The Dna Binding Domains Of Human Fli1 And Runx2 In Complex With 16-Mer Dna Cagaggatgtggcttc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.31 Å Release Date: 2020-11-25 Classification: TRANSCRIPTION |
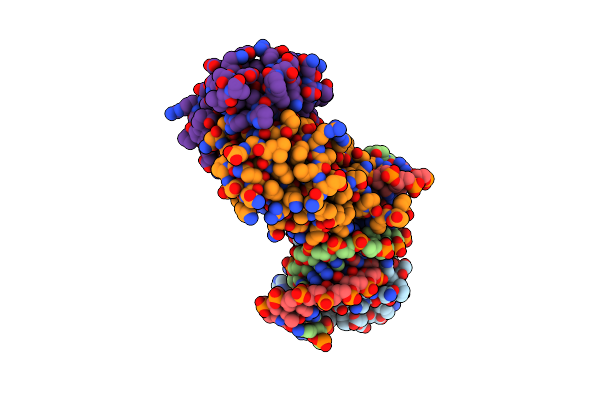 |
Crystal Structure Of The Dna Binding Domain (Dbd) Of Human Fli1 And The Complex Of The Dbd Of Human Runx2 With Core Binding Factor Beta (Cbfb), In Complex With 16Mer Dna Cagaggatgtggcttc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2020-11-25 Classification: TRANSCRIPTION |
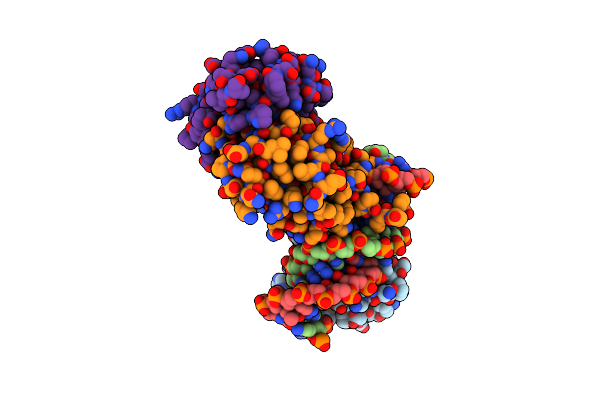 |
Crystal Structure Of The Dna Binding Domains Of Human Transcription Factor Erg, Human Runx2 Bound To Core Binding Factor Beta (Cbfb), In Complex With 16Mer Dna Cagaggatgtggcttc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.25 Å Release Date: 2020-11-25 Classification: TRANSCRIPTION |
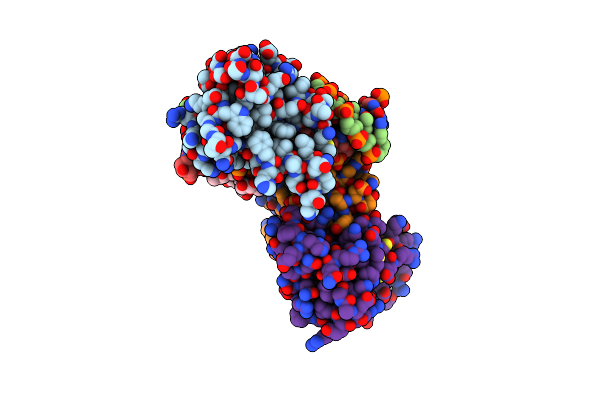 |
Crystal Structure Of The Dna Binding Domains Of Human Transcription Factor Erg, Human Runx2 Bound To Core Binding Factor Beta (Cbfb), And Mithramycin, In Complex With 16Mer Dna Cagaggatgtggcttc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.31 Å Release Date: 2020-11-25 Classification: TRANSCRIPTION Ligands: QWP, MG |
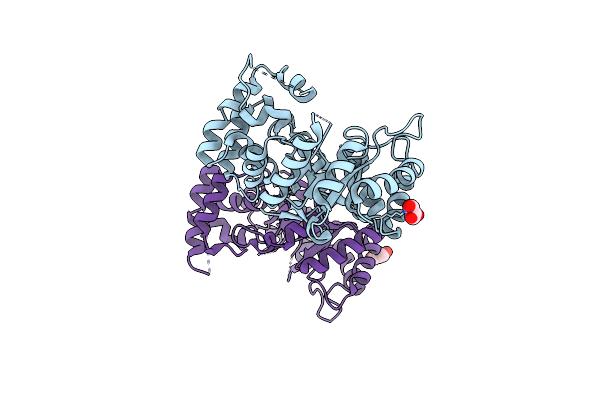 |
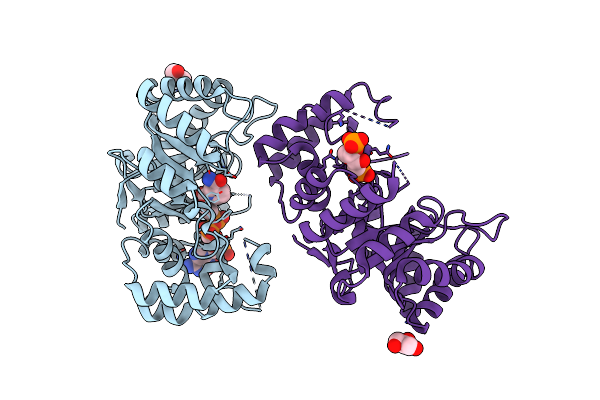
Organism: Streptomyces argillaceus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
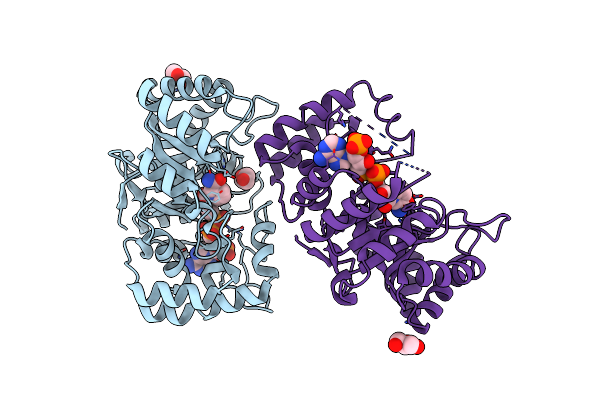
Crystal Structure Of Mithramycin 3-Side Chain Keto-Reductase Mtmw In Complex With Nad+, P422 Form
Organism: Streptomyces argillaceus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: NAP, GOL |
 |
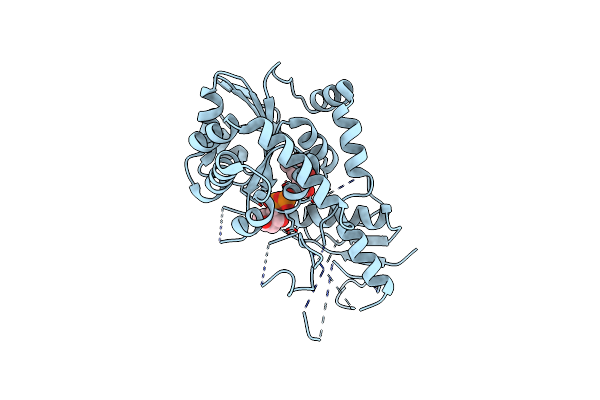
Crystal Structure Of Mithramycin 3-Side Chain Keto-Reductase Mtmw In Complex With Nad+ And Peg
Organism: Streptomyces argillaceus
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: PEG, NAP, GOL |
 |
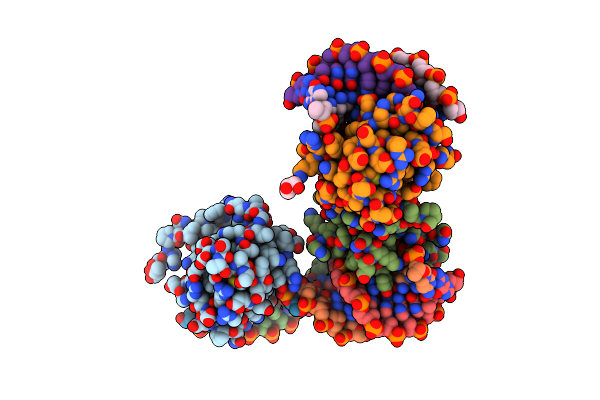
Structural Basis For Earp-Mediated Arginine Glycosylation Of Translation Elongation Factor Ef-P
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-10-04 Classification: TRANSFERASE Ligands: TRH |
 |
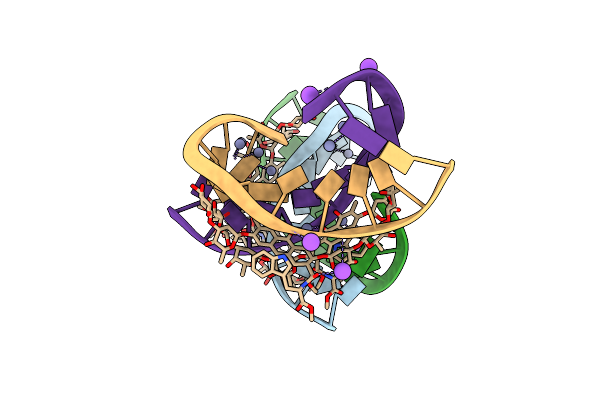
Crystal Structure Of The Dna Binding Domain Of Transcription Factor Fli1 In Complex With An 11-Mer Dna Gaccggaagtg
Organism: Homo sapiens, Endothia gyrosa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-14 Classification: transcription/dna Ligands: PO4, GOL |
 |
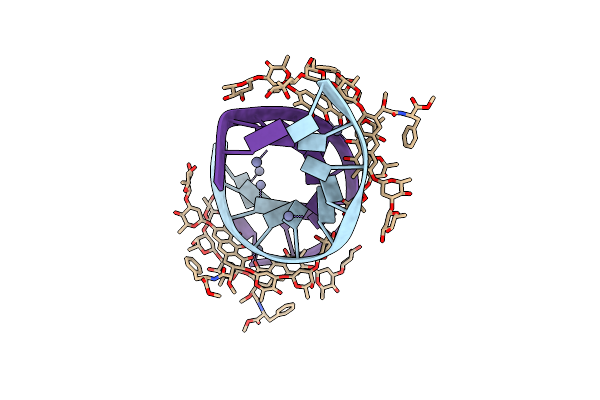
Crystal Structure Of Mithramycin Analogue Mtm Sa-Trp In Complex With A 10-Mer Dna Agaggcctct.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-09-14 Classification: dna/antibiotic Ligands: ZN, NA, 6O6 |
 |
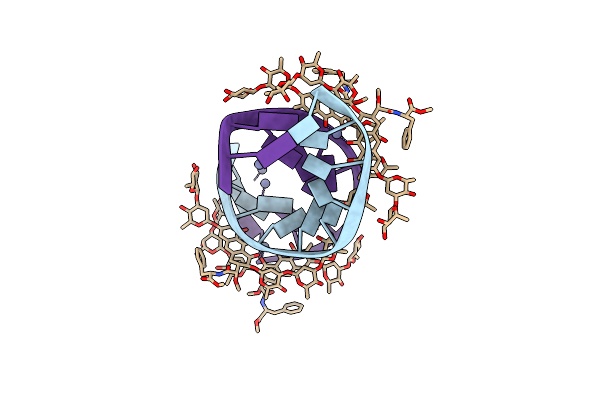
Crystal Structure Of Mithramycin Analogue Mtm Sa-Phe In Complex With A 10-Mer Dna Agggtaccct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-09-14 Classification: dna/antibiotic Ligands: 6O7, ZN |
 |
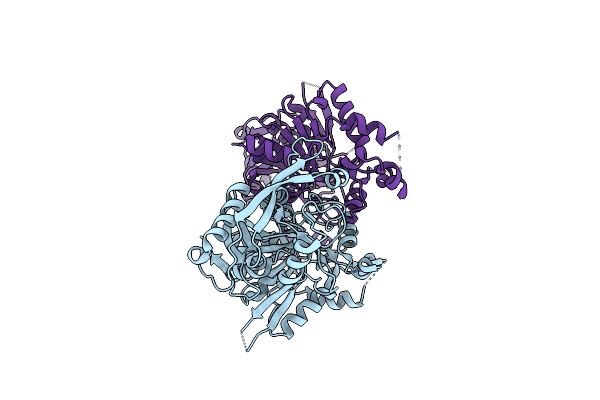
Crystal Structure Of Mithramycin Analogue Mtm Sa-Phe In Complex With A 10-Mer Dna Agggatccct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-14 Classification: DNA/antibiotic Ligands: 6O7, ZN |
 |
Organism: Cryptococcus neoformans var. grubii serotype a (strain h99 / atcc 208821 / cbs 10515 / fgsc 9487)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-08-24 Classification: LIGASE |
 |
Adenylosuccinate Synthetase From Cryptococcus Neoformans Complexed With Gdp And Imp
Organism: Cryptococcus neoformans var. grubii serotype a (strain h99 / atcc 208821 / cbs 10515 / fgsc 9487)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-08-24 Classification: LIGASE Ligands: GDP, IMP |
 |
Structural And Functional Studies Of Bexe: Insights Into Oxidation During Be-7585A Biosynthesis
Organism: Amycolatopsis orientalis subsp. vinearia
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-12-02 Classification: OXIDOREDUCTASE Ligands: SO4, FAD |
 |
Organism: Streptomyces argillaceus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-02-04 Classification: TRANSFERASE Ligands: ZN, SAH, ACT, CL |
 |
Organism: Streptomyces cyanogenus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2015-01-28 Classification: TRANSFERASE |