Search Count: 46
 |
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN |
 |
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of The Nipah Virus Polymerase Containing The Connecting Domain
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN, TRANSFERASE |
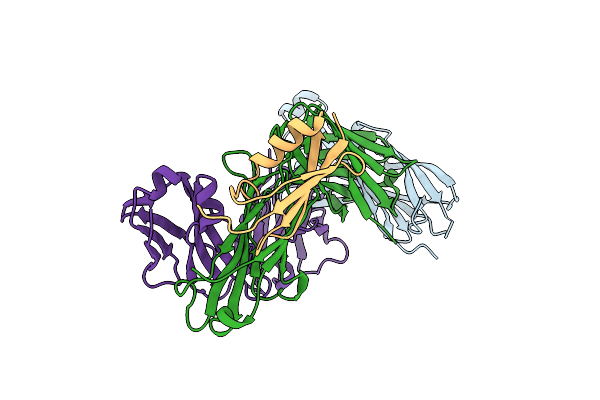 |
Organism: Homo sapiens, Streptococcus sp. 'group g'
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-05 Classification: IMMUNE SYSTEM/Chaperone |
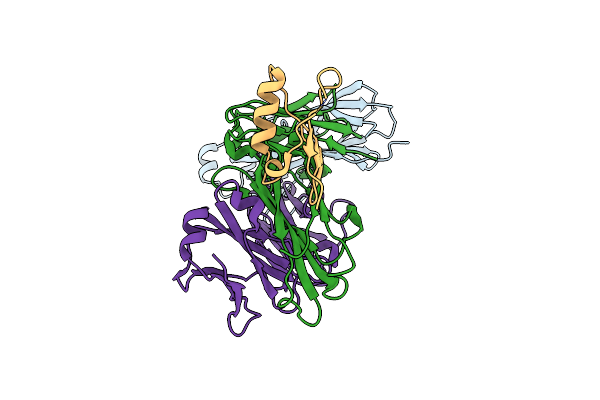 |
Organism: Homo sapiens, Streptococcus sp. 'group g'
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-02-05 Classification: IMMUNE SYSTEM |
 |
Organism: Alcanivorax borkumensis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: VVA |
 |
Organism: Alkalilimnicola ehrlichii (strain atcc baa-1101 / dsm 17681 / mlhe-1), Ruegeria pomeroyi
Method: X-RAY DIFFRACTION Resolution:4.19 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: PE4, D12, OCT, 12P, 1PE |
 |
Organism: Homo sapiens, Alkalilimnicola ehrlichii mlhe-1, Ruegeria pomeroyi dss-3
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Alkalilimnicola ehrlichii (strain atcc baa-1101 / dsm 17681 / mlhe-1), Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN Ligands: 4NB, LHG, OCT, PE4, LNK, 1PE, D12, R16, BHC, GOL |
 |
Organism: Ruegeria pomeroyi
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Alcanivorax borkumensis (strain atcc 700651 / dsm 11573 / ncimb 13689 / sk2)
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN Ligands: ACT, NA, MG |
 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BA, DGA |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BA |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Crystal Structure Of An Archaeal Cnnm, Mtcorb, With C-Terminal Deletion In Complex With Mg2+-Atp
Organism: Methanoculleus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.26 Å Release Date: 2021-06-16 Classification: MEMBRANE PROTEIN Ligands: MG, SO4, UMQ, ATP, 1PE |
 |
Crystal Structure Of An Archaeal Cnnm, Mtcorb, R235L Mutant With C-Terminal Deletion
Organism: Methanoculleus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2021-06-16 Classification: MEMBRANE PROTEIN |

