Search Count: 38
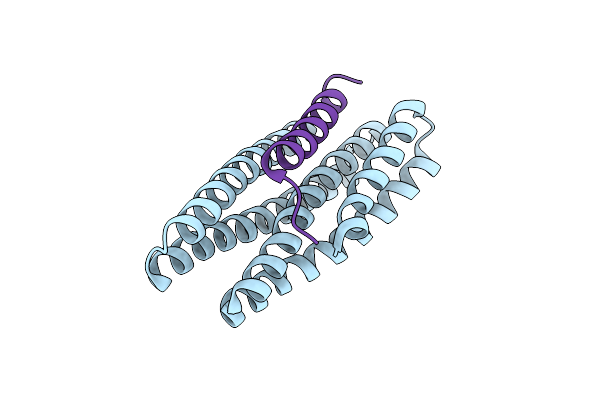 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
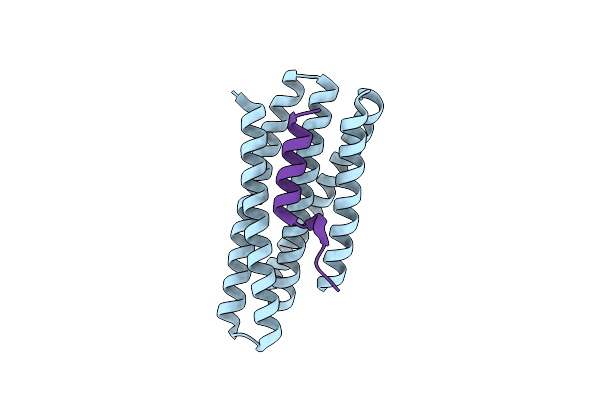 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
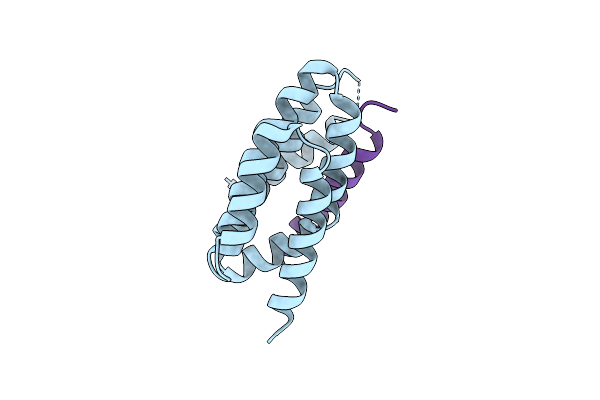 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
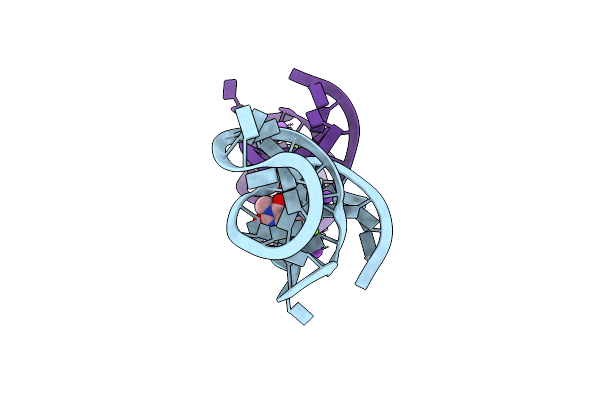 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-11-30 Classification: RNA Ligands: TEP, MG, NA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-11-30 Classification: RNA/IMMUNE SYSTEM Ligands: K, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-11-30 Classification: RNA Ligands: QB3, NA, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-11-30 Classification: RNA Ligands: QAX, MG, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-11-30 Classification: RNA Ligands: QIJ, MG, CA, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-11-30 Classification: RNA Ligands: QEU |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-11-30 Classification: RNA/IMMUNE SYSTEM |
 |
Non-Receptor Protein Tyrosine Phosphatase Shp2 In Complex With Allosteric Inhibitor Rmc-4550
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-01 Classification: SIGNALING PROTEIN Ligands: 4Q4, CL |
 |
1.80A Resolution Structure Of Independent Phosphoglycerate Mutase From C. Elegans In Complex With A Macrocyclic Peptide Inhibitor (Ce-1 Nhoh)
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-07 Classification: ISOMERASE Ligands: ZN, NA, PG4 |
 |
2.10A Resolution Structure Of Independent Phosphoglycerate Mutase From C. Elegans In Complex With A Macrocyclic Peptide Inhibitor (Ce-2 Y7F)
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-07 Classification: ISOMERASE Ligands: CL, NA, ZN |
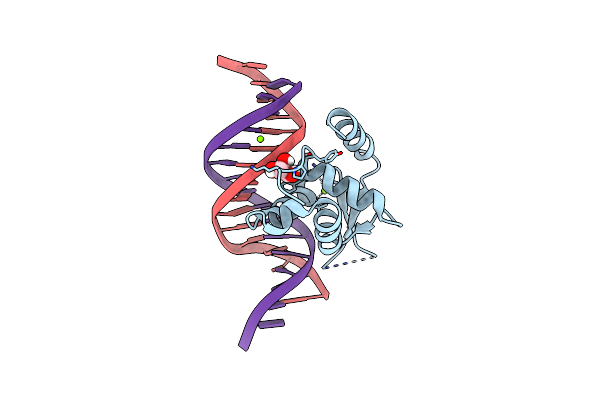 |
Crystal Structure Of The Human Foxn3 Dna Binding Domain In Complex With A Forkhead Dna Sequence
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-02-27 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, GOL |
 |
Crystal Structure Of The Human Foxn3 Dna Binding Domain In Complex With A Forkhead-Like (Fhl) Dna Sequence
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-27 Classification: DNA BINDING PROTEIN/DNA Ligands: MG |
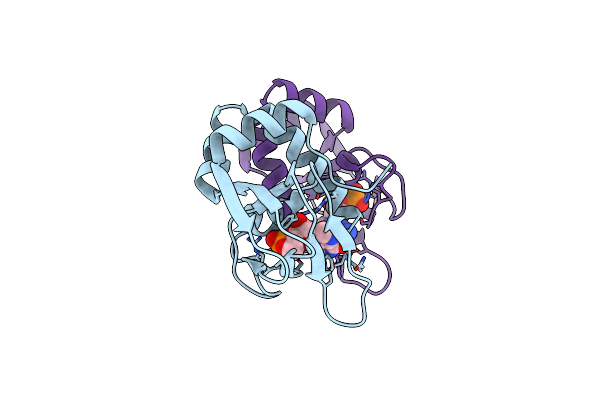 |
A Cyclic-Gmp-Dependent Signalling Pathway Regulates Bacterial Phytopathogenesis
Organism: Xanthomonas campestris pv. campestris (strain b100)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-05-24 Classification: BIOSYNTHETIC PROTEIN Ligands: PCG |
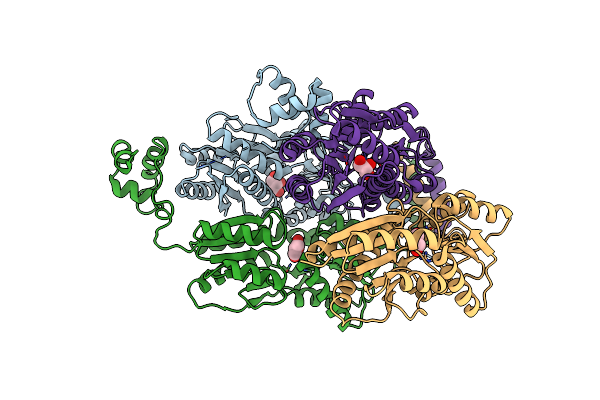 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2015-12-23 Classification: TRANSCRIPTION Ligands: GOL |
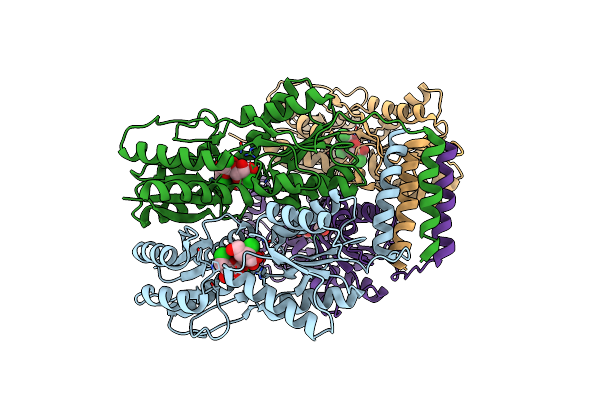 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-12-23 Classification: TRANSCRIPTION |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-04-08 Classification: DNA BINDING PROTEIN Ligands: SO4, 480, GOL |

