Search Count: 55
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Structure Of Candida Albicans 80S Ribosome In Complex With Mefloquine (Non-Rotated State)
Organism: Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: RIBOSOME Ligands: ZN, YMZ, SPK |
 |
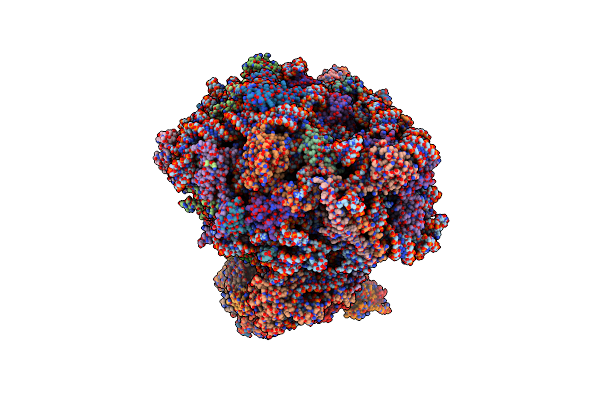
The Structure Of The Candida Albicans Ribosome With Trna-Fmet, Mrna, And Compounds (Gen And Mfq) Shows Strong Density For The A Site Trna
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: RIBOSOME Ligands: SPK, GET, ZN |
 |
The Structure Of The Candida Albicans Ribosome With Trna-Fmet, Mrna, And Compounds (Gen And Mfq) With Strong Density For The P-Site Trna
Organism: Escherichia coli, Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: RIBOSOME Ligands: SPK, GET, ZN, YMZ |
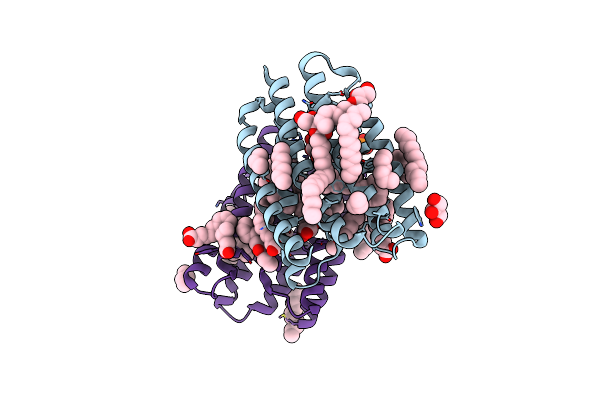 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
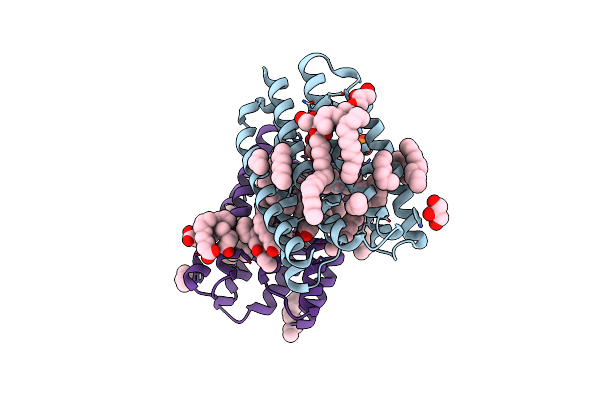 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
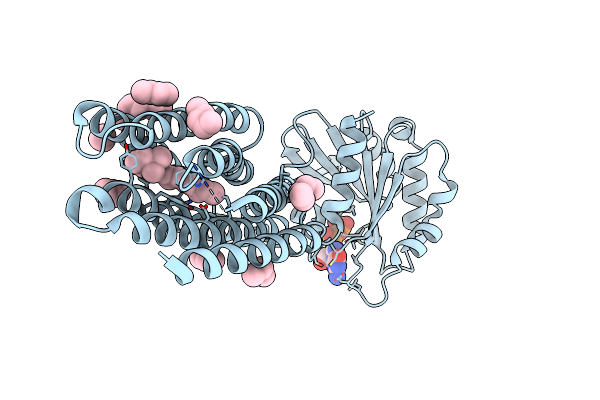 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4, NA |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The L State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, NA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: OLA, OLC, LFA, PO4, NA |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.6 In The Absence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, PO4 |

