Search Count: 18
 |
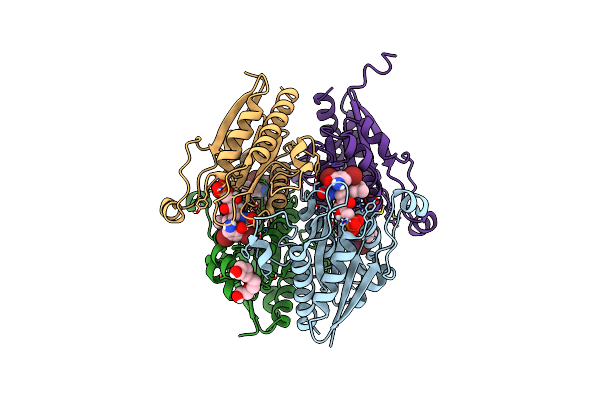
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-08-03 Classification: FLAVOPROTEIN Ligands: CL |
 |
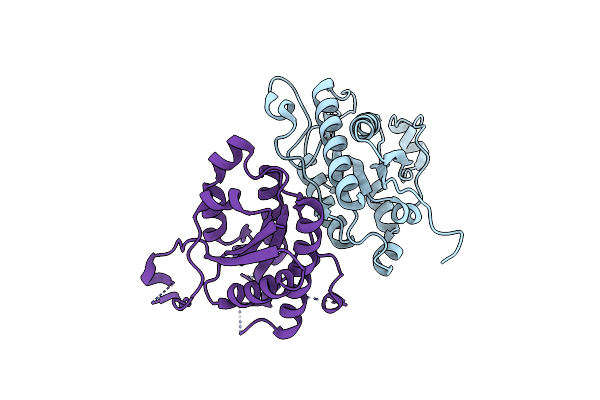
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-08-03 Classification: FLAVOPROTEIN Ligands: 8YX, FMN, PG4 |
 |
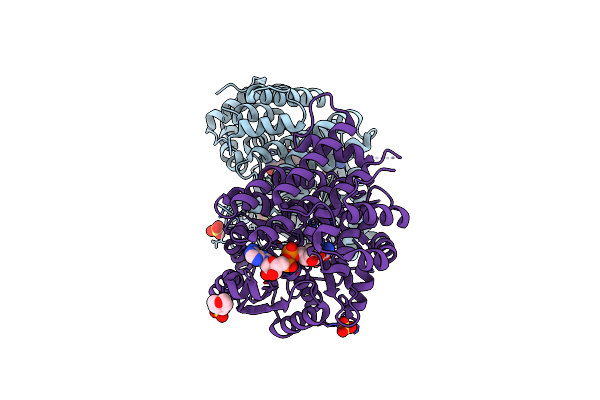
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-08-03 Classification: FLAVOPROTEIN Ligands: CL |
 |
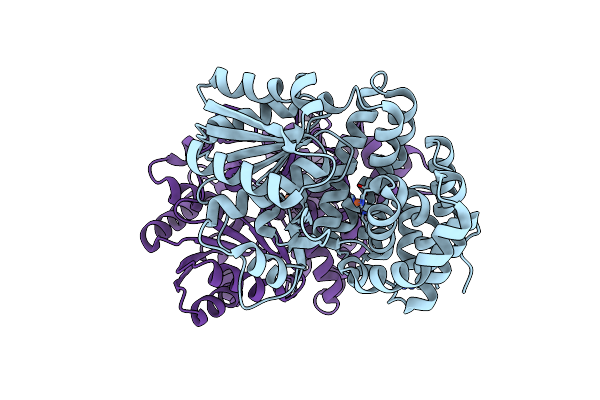
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, EDO, FE, MES |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Cryo-Em Structure Of Aldehyde-Alcohol Dehydrogenase Reveals A High-Order Helical Architecture Critical For Its Activity
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: HYDROLASE |
 |
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-17 Classification: LIPID BINDING PROTEIN Ligands: VCA, EDO, TAM |
 |
Solution Structure Of As-P18 Reveals That Nematode Fatty Acid Binding Proteins Exhibit Unusual Structural Features
Organism: Ascaris suum
Method: SOLUTION NMR Release Date: 2019-07-17 Classification: LIPID BINDING PROTEIN Ligands: OLA |
 |
Diversity In The Structures And Ligand Binding Sites Among The Fatty Acid And Retinol Binding Proteins Of Nematodes Revealed By Na-Far-1 From Necator Americanus
Organism: Necator americanus
Method: SOLUTION NMR Release Date: 2015-09-16 Classification: RETINOL-BINDING PROTEIN |
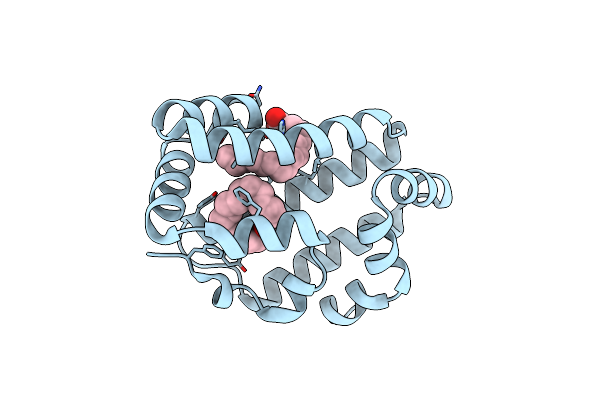 |
Organism: Necator americanus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2015-09-16 Classification: retinol-binding protein Ligands: PLM |
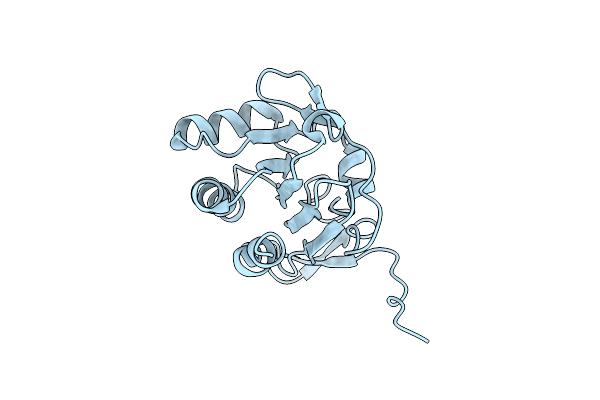 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2012-05-02 Classification: OXIDOREDUCTASE |
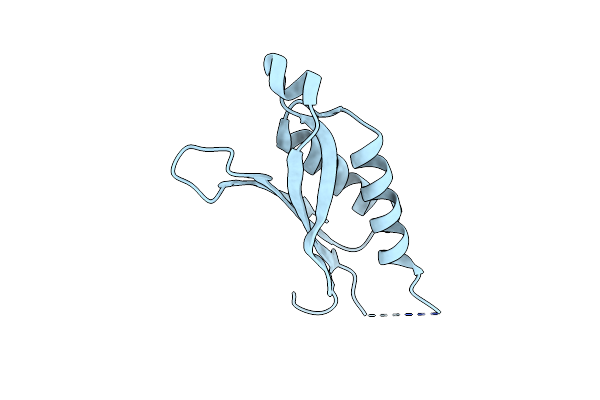 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-03-21 Classification: ISOMERASE |
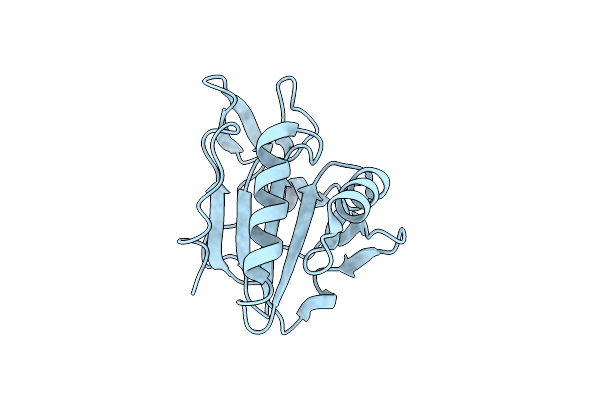 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2012-03-14 Classification: OXIDOREDUCTASE |
 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2012-03-14 Classification: OXIDOREDUCTASE |
 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2012-03-14 Classification: OXIDOREDUCTASE |
 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-08-10 Classification: OXIDOREDUCTASE |
 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-29 Classification: OXIDOREDUCTASE Ligands: DTU |

