Search Count: 12
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-01-31 Classification: LIPID TRANSPORT |
 |
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-01-31 Classification: LIPID TRANSPORT |
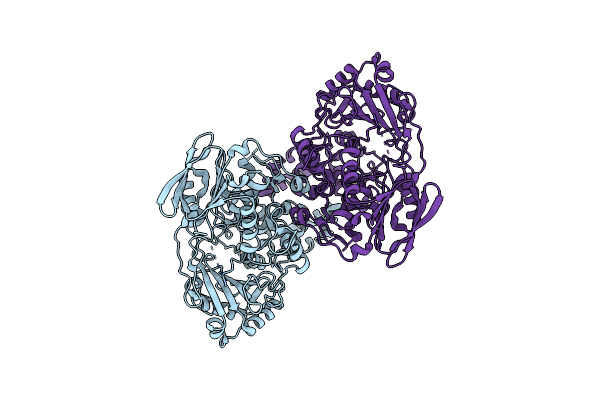 |
Cryo-Em Reconstruction Of Staphylococcus Aureus Oleate Hydratase (Ohya) Dimer With An Ordered C-Terminal Membrane-Association Domain
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: HYDROLASE |
 |
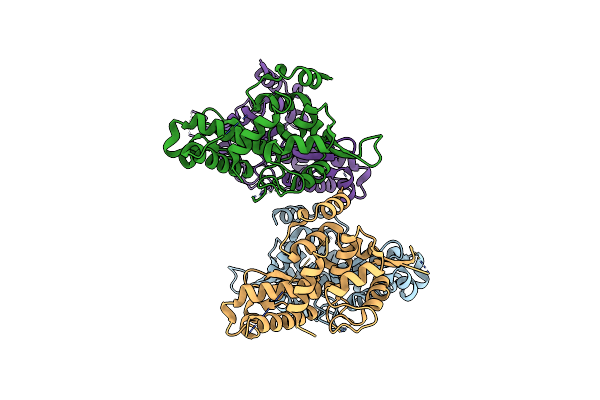
Cryo-Em Reconstruction Of Staphylococcus Aureus Oleate Hydratase (Ohya) Dimer With A Disordered C-Terminal Membrane-Association Domain
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: HYDROLASE |
 |
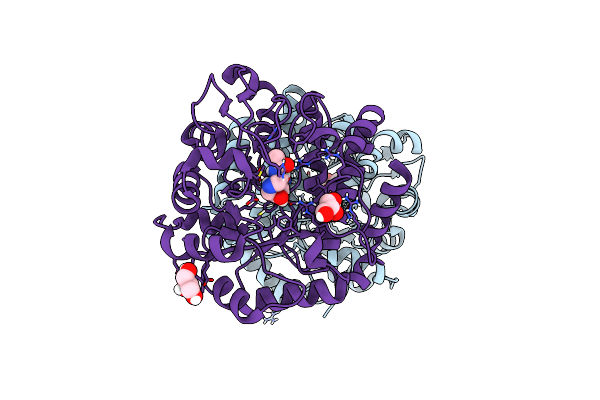
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-10-14 Classification: LIGASE |
 |
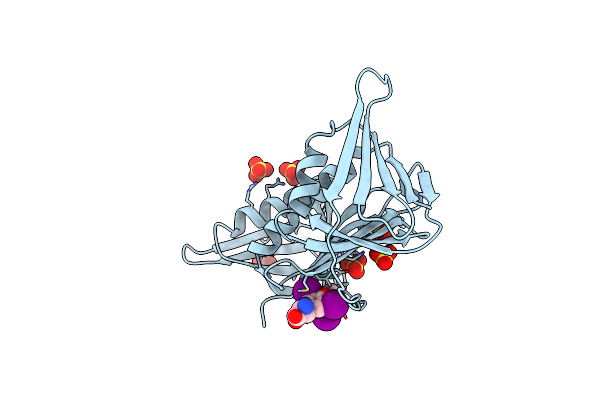
Crystal Structure Of Burkholderia Cenocepacia Family 3 Glycoside Hydrolase (Nagz) Bound To (3S,4R,5R,6S)-3-Acetamido-4,5,6-Trihydroxyazepane
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2CZ, GOL |
 |
Structure Of An Rsp5Xubxsna3 Complex: Mechanism Of Ubiquitin Ligation And Lysine Prioritization By A Hect E3
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-08-14 Classification: Ligase/protein binding |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-03-14 Classification: DNA BINDING PROTEIN Ligands: T3, SO4, CL |
 |
The Nmr Solution Structure Of The 3Fe Ferredoxin Ii From Desulfovibrio Gigas, 15 Structures
Organism: Desulfovibrio gigas
Method: SOLUTION NMR Release Date: 1999-09-02 Classification: ELECTRON TRANSPORT Ligands: F3S |

