Search Count: 19
 |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of A Selenium-Derivatized Complex Of The Bacterial Cellulose Secretion Regulators Bcsr And Bcsq, Crystallized In The Presence Of Appcp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: MG, ATP |
 |
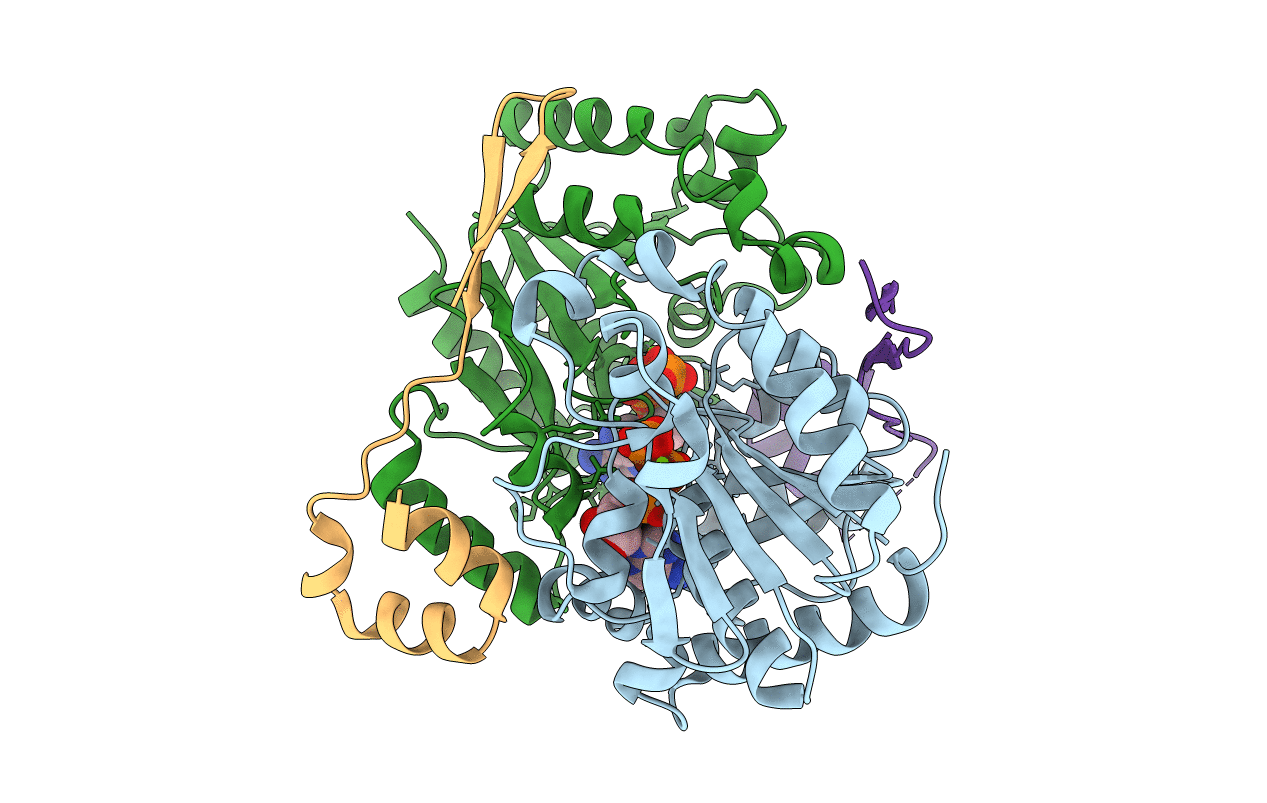
Crystal Structure Of A Selenium-Derivatized Complex Of The Bacterial Cellulose Secretion Regulators Bcsr And Bcsq, Crystallized In The Presence Of Adp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: MG, ATP |
 |
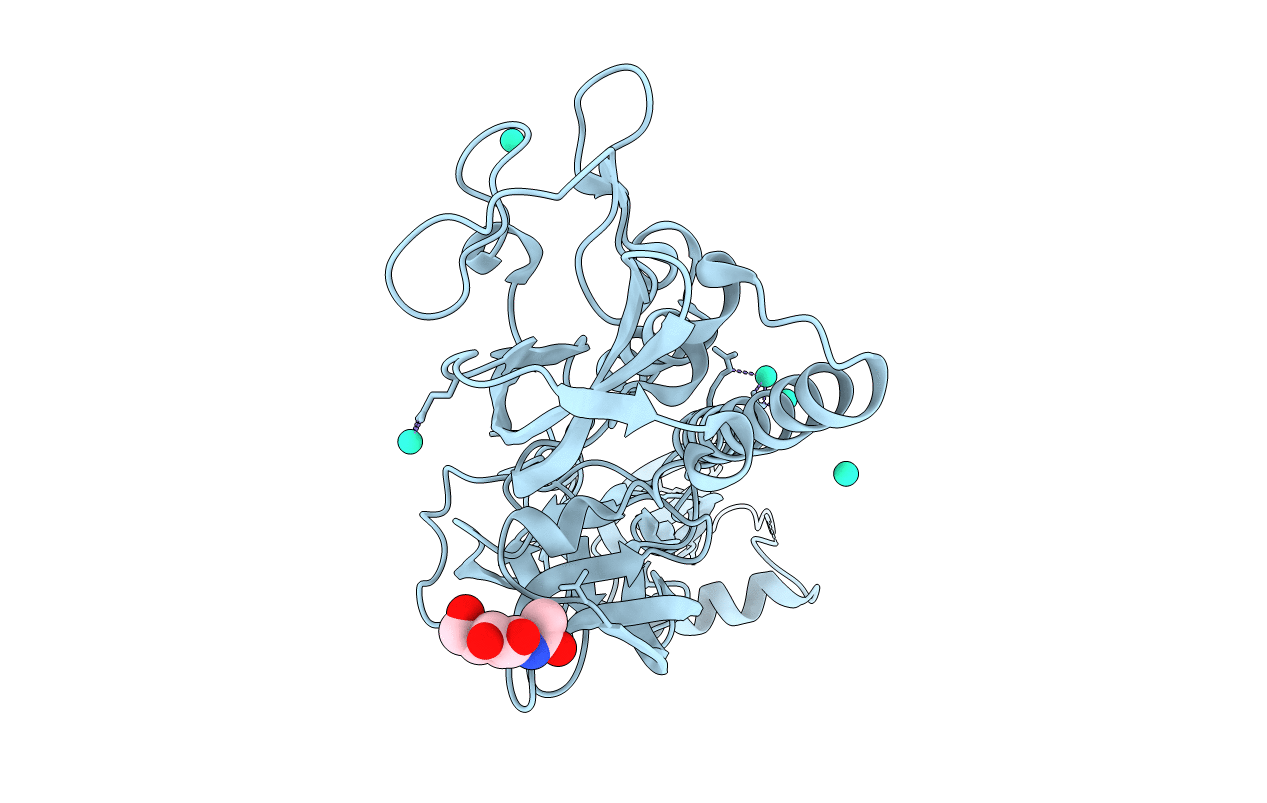
Crystal Structure Of A Native Bcsrq Complex Purified And Crystallized In The Absence Of Nucleotide
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: MG, ATP |
 |
Orthorhombic Crystal Structure Of A Native Bcsrq Complex Crystallized In The Presence Of Adp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: MG, ATP |
 |
Crystal Structure Of A Native Bcse (217-523) - Bcsr-Bcsq (R156E Mutant) Complex With C-Di-Gmp And Atp Bound
Organism: Escherichia coli, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: MG, ATP, C2E, GOL |
 |
Crystal Structure Of A Native Bcse (349-523) Rq Complex With C-Di-Gmp And Atp Bound
Organism: Escherichia coli, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: ATP, MG, C2E |
 |
Cryo-Em Structure Of A Bcsb Pentamer In The Context Of An Assembled Bcs Macrocomplex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: SIGNALING PROTEIN |
 |
Organism: Mokola virus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2020-02-19 Classification: VIRAL PROTEIN Ligands: NAG, TB |
 |
Structure Of Pragmin Pseudo-Kinase Reveals A Dimerization Mechanism To Regulate Protein Tyrosine Phosphorylation And Nuclear Transcription
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2018-01-31 Classification: CELL INVASION Ligands: SO4 |
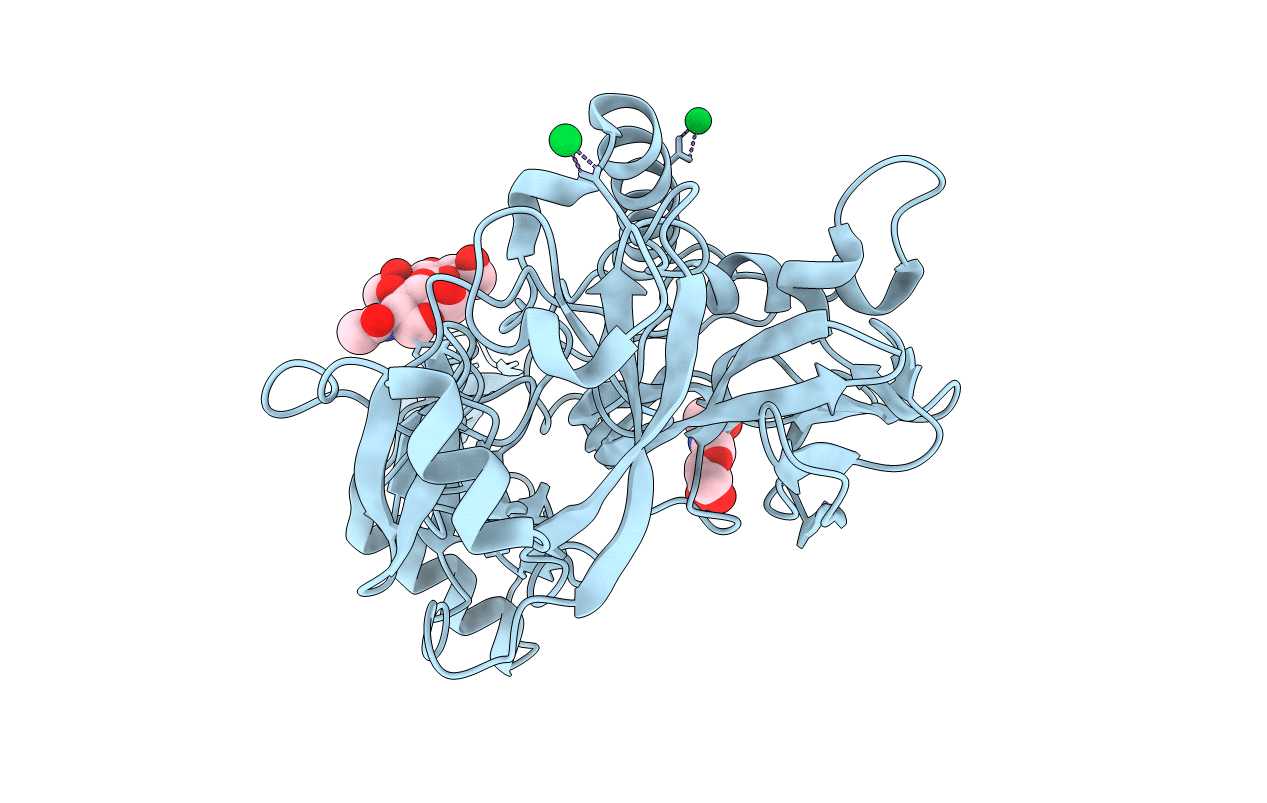 |
Organism: Vesicular stomatitis indiana virus (strain mudd-summers)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-02 Classification: VIRAL PROTEIN Ligands: NAG, YB |
 |
Crystal Structure Of Vsv-Indiana (Mudd-Summers Strain) Glycoprotein Under Its Acidic Conformation
Organism: Vesicular stomatitis indiana virus (strain mudd-summers)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-02-24 Classification: MEMBRANE PROTEIN Ligands: PE4, CA |
 |
Crystal Structure Of Fragment 1600-1733 Of Hsv1 Ul36 In The Presence Of 1M Potassium Iodide
Organism: Human herpesvirus 1(type 1 / strain 17)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-02-18 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Herpes simplex virus (type 1 / strain 17)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-02-18 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-10-06 Classification: NUCLEIC ACID BINDING PROTEIN Ligands: EDO, MG |
 |
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-10-06 Classification: NUCLEIC ACID BINDING PROTEIN Ligands: MG, EDO |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-10-27 Classification: NUCLEIC ACID BINDING PROTEIN Ligands: CL, EDO |
 |
Structure Of The C-Terminal Domain Of The Polymerase Cofactor Of Rabies Virus: Insights In Function And Evolution.
Organism: rabies virus cvs-11
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-10-15 Classification: TRANSFERASE Ligands: GOL |

