Search Count: 46
 |
Crystal Structure Of Methanopyrus Kandleri Malate Dehydrogenase Mutant 4 At Room Temperature
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, GOL, CL, NA |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, NA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: TRANSPORT PROTEIN Ligands: PEX |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: TRANSPORT PROTEIN Ligands: PEX |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: TRANSPORT PROTEIN Ligands: PEX |
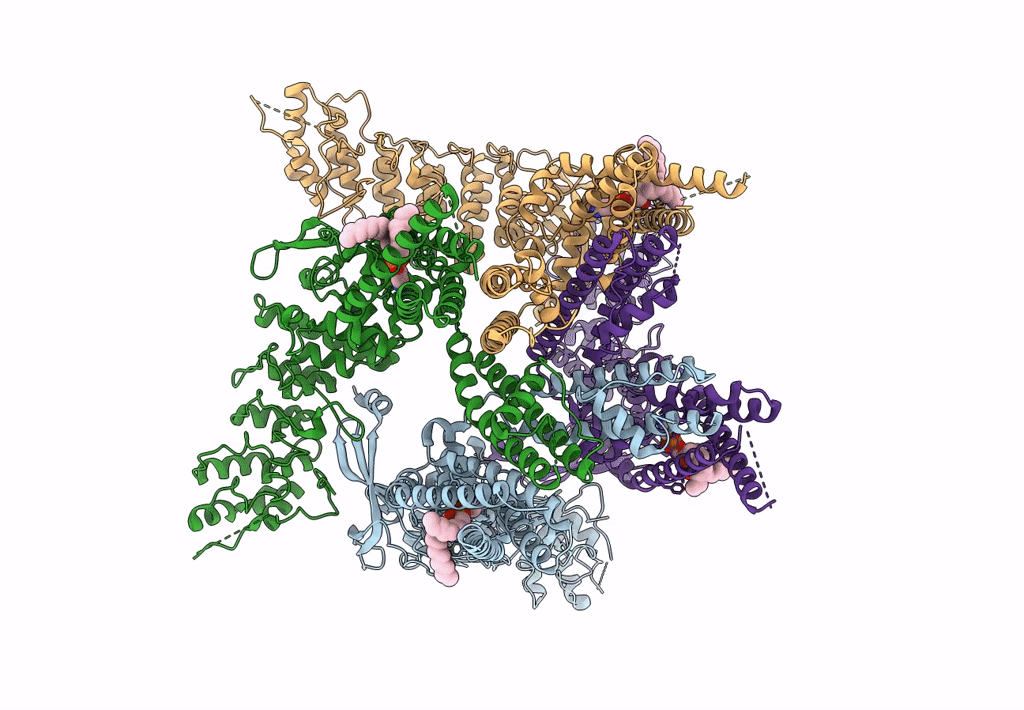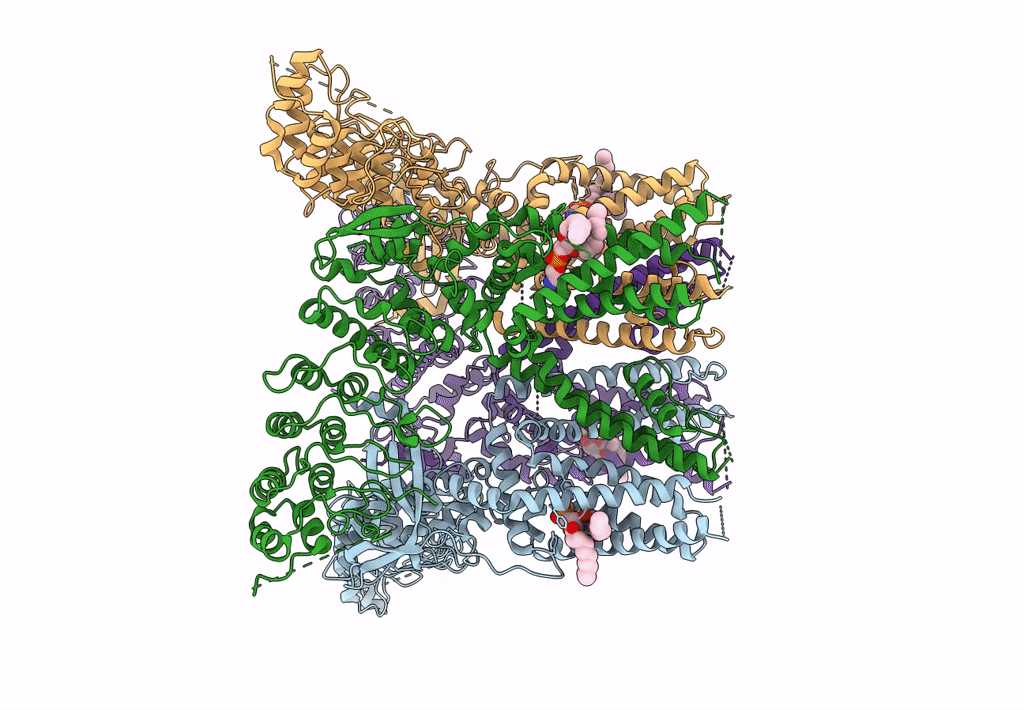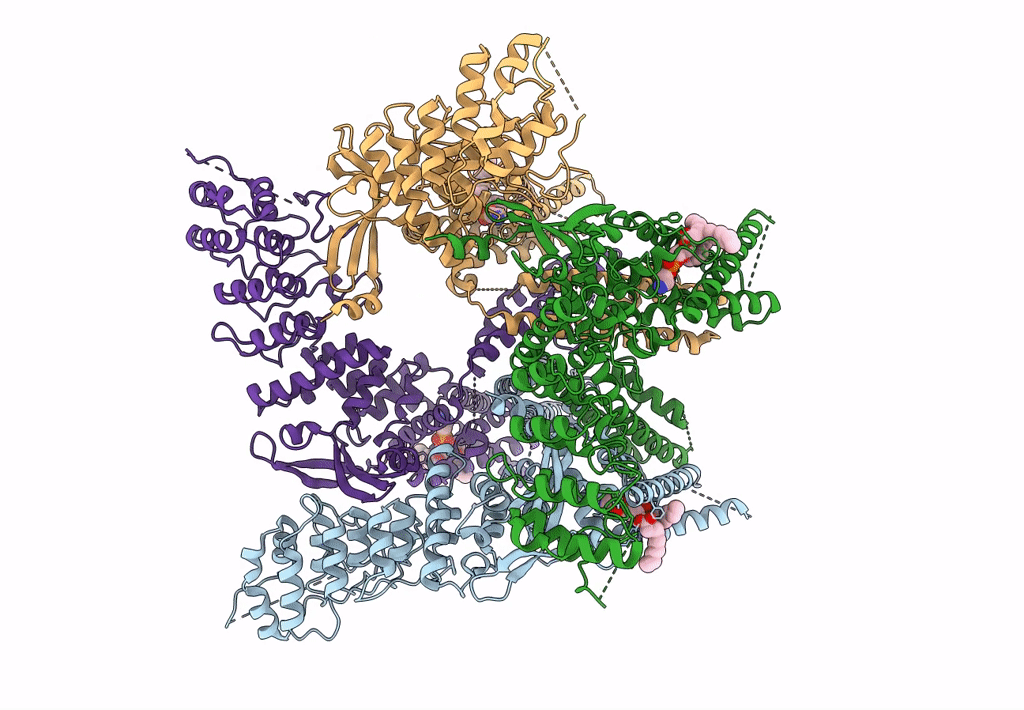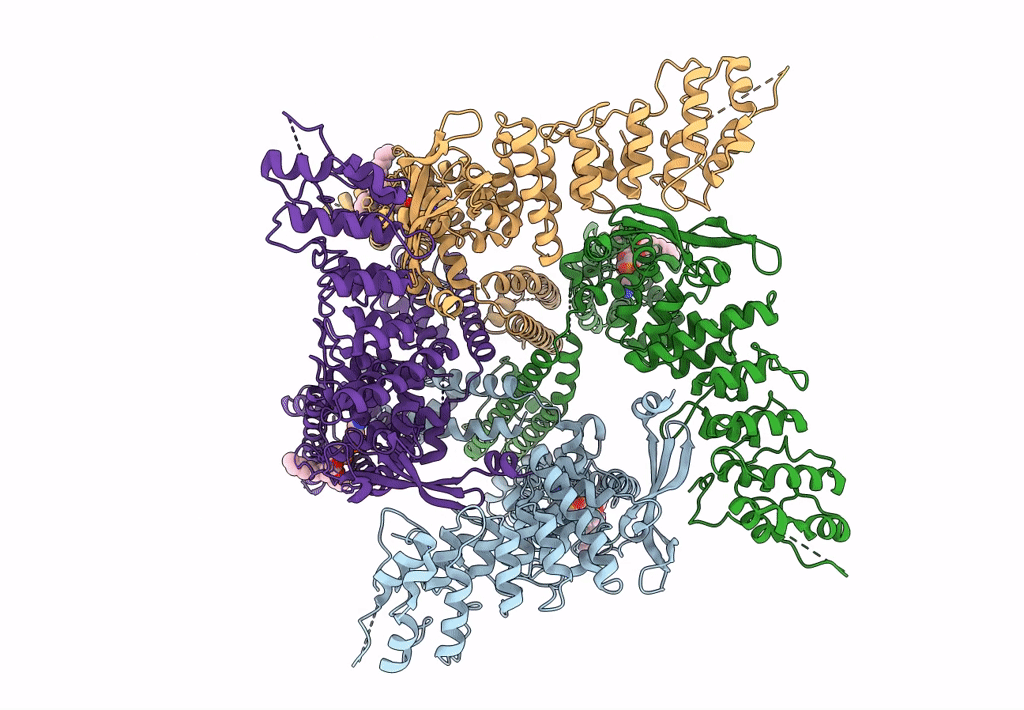 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: TRANSPORT PROTEIN Ligands: PEX |
 |
Microtubule Decorated With Tubulin Oligomers In Presence Of Apc C-Terminal Domain. (Here Only Map Corresponding To The 13-Pf Microtubule Is Represented)
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: CELL CYCLE Ligands: TA1, GDP, MG, GTP |
 |
Crystal Structure Of Malate Dehydrogenase From Haloarcula Marismortui With Potassium And Chloride Ions
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: OXIDOREDUCTASE Ligands: CL, K |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-09 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Synergistic Stabilization Of A Double Mutant In Ci2 From An In-Cell Library Screen
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2020-12-09 Classification: PROTEIN BINDING |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-12-09 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-12-09 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Crystal Structure Of Ignicoccus Islandicus Malate Dehydrogenase Co-Crystallized With 10 Mm Tb-Xo4
Organism: Ignicoccus islandicus dsm 13165
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:1.89 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: 7MT, TB, CL |
 |
Structure Of A Psychrophilic Cca-Adding Enzyme At Room Temperature In Chipx Microfluidic Device
Organism: Planococcus halocryophilus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-05-29 Classification: RNA BINDING PROTEIN |
 |
Structure Of A Nonameric Rna Duplex At Room Temperature In Chipx Microfluidic Device
Organism: Rattus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-01 Classification: RNA Ligands: SO4 |
 |
Structure Of Protease1 From Pyrococcus Horikoshii At Room Temperature In Chipx Microfluidic Device
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-05-01 Classification: HYDROLASE |
 |
Structure Of A Psychrophilic Cca-Adding Enzyme In Complex With Cmpcpp At Room Temperature In Chipx Microfluidic Device
Organism: Planococcus halocryophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-05-01 Classification: RNA BINDING PROTEIN Ligands: 2TM |
 |
Room Temperature Structure Of Lipase From T. Lanuginosa At 2.5 A Resolution In Chipx Microfluidic Device
Organism: Thermomyces lanuginosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: NAG, PO4, MG |

