Search Count: 18
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalently Bound Inhibitor Fp237 (Compound 8P In Publication)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2025-01-29 Classification: VIRAL PROTEIN Ligands: A1IPK, DMS, PEG, EDO, NA, CL |
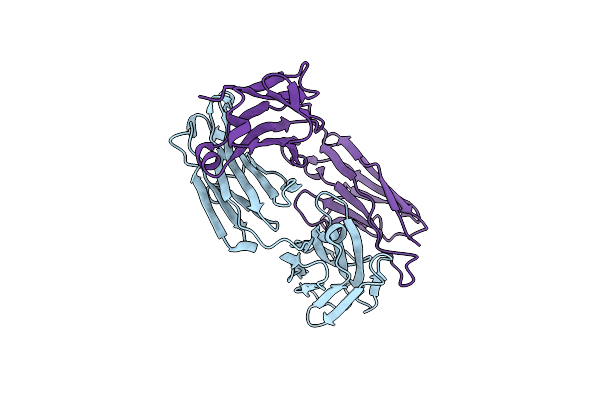 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-03-27 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Organism: Blautia obeum
Method: X-RAY DIFFRACTION Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: SAM, BTB, Q46, SF4, DTB, FE, CL |
 |
Organism: Veillonella parvula
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: Q46, SAM, SF4, DTB, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2023-10-25 Classification: VIRAL PROTEIN/INHIBITOR Ligands: LKR |
 |
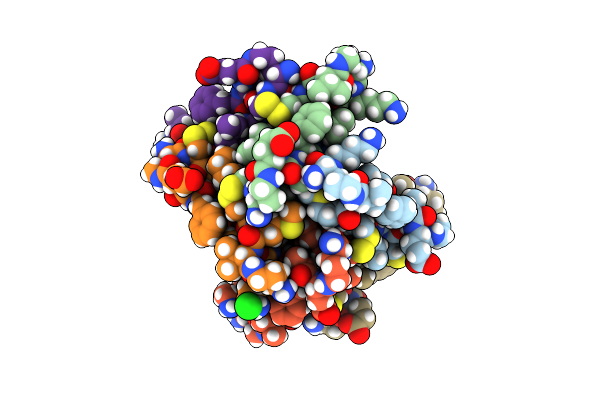
Covalently Stabilized Triangular Trimer Derived From Abeta16-36 With P-Iodo-Phenylalanine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-05-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-05-31 Classification: DE NOVO PROTEIN Ligands: MPD, CL |
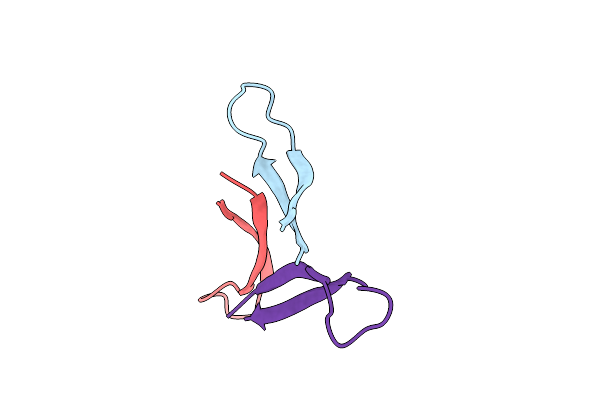 |
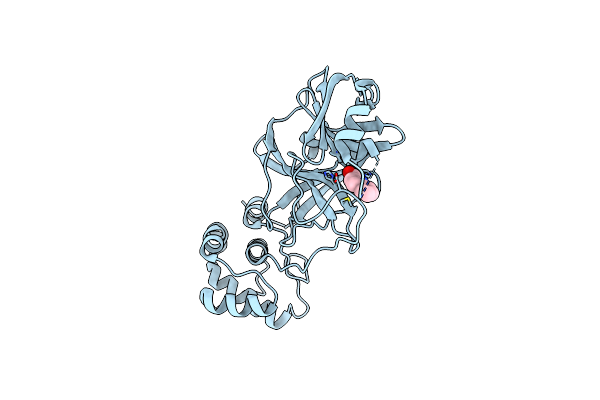
Covalently Stabilized Triangular Trimer Composed Of Abeta17-36 Beta-Hairpins
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-15 Classification: DE NOVO PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-06 Classification: VIRAL PROTEIN/INHIBITOR Ligands: QNC |
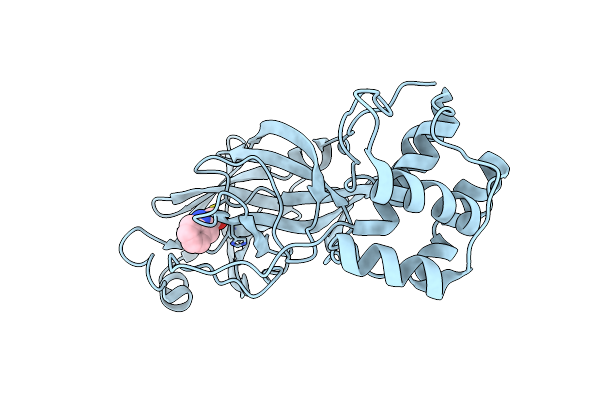 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-07-06 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 9FF |
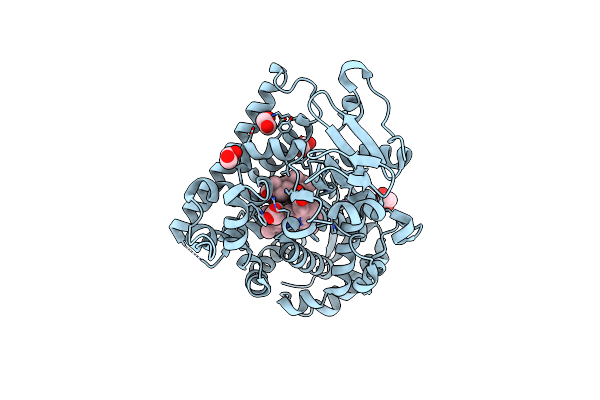 |
Organism: Bacillus megaterium (strain atcc 14581 / dsm 32 / jcm 2506 / nbrc 15308 / ncimb 9376 / nctc 10342 / vkm b-512)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2020-04-29 Classification: OXIDOREDUCTASE Ligands: TES, IMD, EDO, HEM |
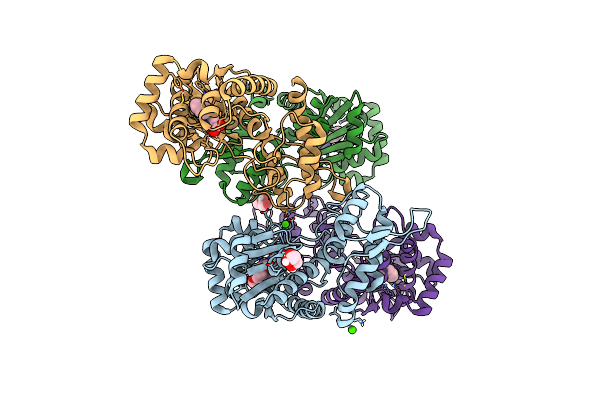 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-01-16 Classification: LIGASE Ligands: 96Z, GOL, CA, PG4 |
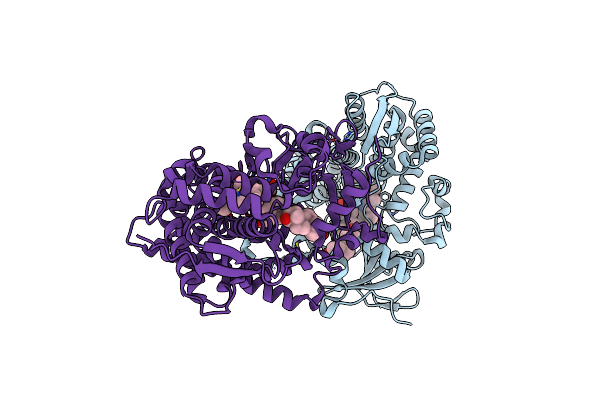 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-08-01 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, TES, CL |
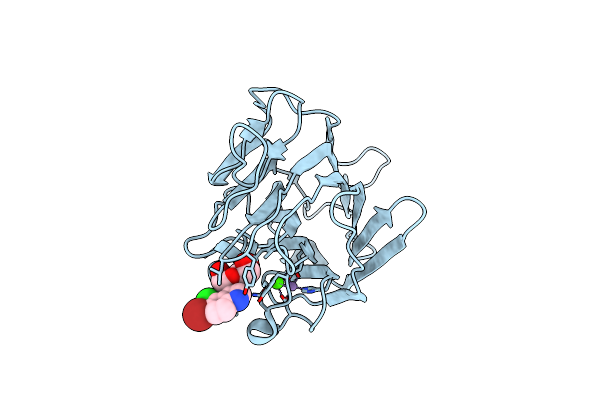 |
Crystal Structure Of Lectin From Dioclea Lasiophylla Seeds (Dlyl) Complexed With X-Man
Organism: Dioclea lasiophylla
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-03-07 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, XMM |
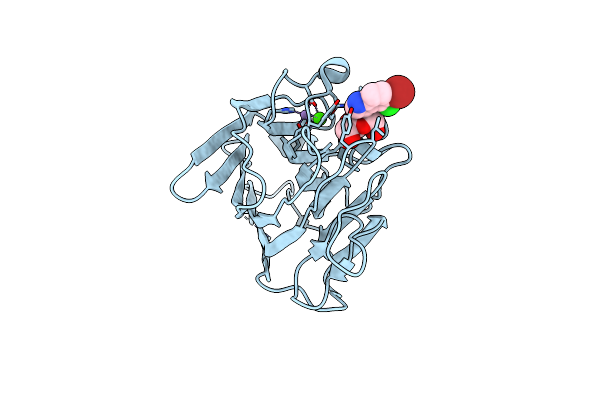 |
Organism: Dioclea lasiocarpa
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-10-04 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA, XMM |
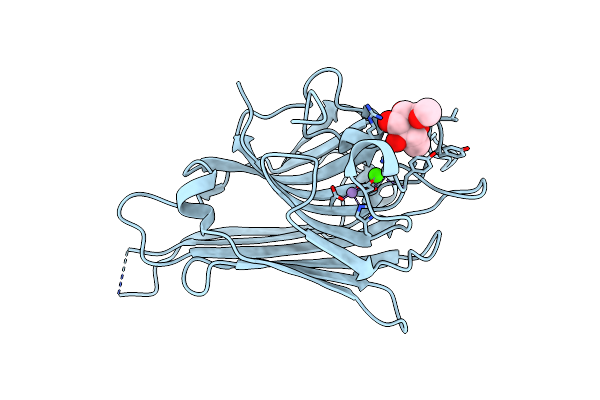 |
Crystal Structure Of Native Lectin From Canavalia Bonariensis Seeds (Cabo) Complexed With Alpha-Methyl-D-Mannoside
Organism: Canavalia bonariensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-23 Classification: SUGAR BINDING PROTEIN Ligands: MMA, MN, CA |
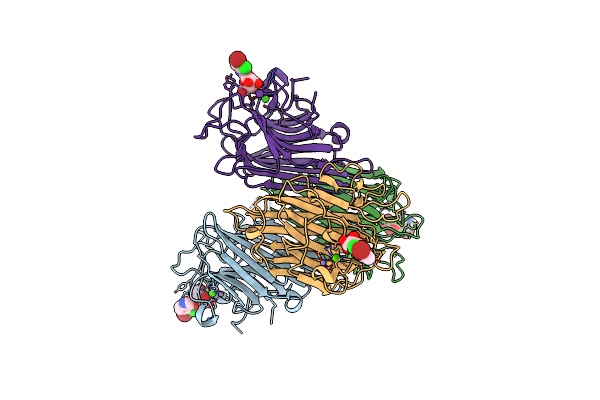 |
Crystal Structure Of Dioclea Reflexa Seed Lectin (Drfl) In Complex With X-Man
Organism: Dioclea reflexa
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2017-02-15 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, XMM |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-30 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, 71E, ACP |

