Search Count: 24
 |
Organism: Didymium iridis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-22 Classification: RNA Ligands: MES, MG, NA |
 |
Organism: Gymnema sylvestre
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-08-08 Classification: PLANT PROTEIN Ligands: NI |
 |
Crystal Structure Of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M In Complex With Sucrose
Organism: Leuconostoc citreum
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2017-11-29 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Leuconostoc citreum
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2017-11-01 Classification: TRANSFERASE Ligands: CA, PR |
 |
Crystal Structure Of Leuconostoc Citreum Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-M
Organism: Leuconostoc citreum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: CA, GOL |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2016-03-23 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2016-02-10 Classification: CELL CYCLE Ligands: GOL, MG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2016-01-27 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-01-20 Classification: REPLICATION |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.60 Angstrom In Complex With N-Acetylglucosamine And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, NDG, GOL, K, EDO |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.85 Angstrom From Unknown Human Gut Bacteria (Uhgb_Mp)
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, GOL, K, EDO, PGE |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.60 Angstrom In Complex With Beta-D-Mannopyranose And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, BMA, K, EDO, PGE |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.76 Angstrom From Unknown Human Gut Bacteria (Uhgb_Mp) In Complex With N-Acetyl-D-Glucosamine, Beta-D-Mannopyranose And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, BMA, NDG, PGE, EDO, K, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-03-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Didymium iridis
Method: X-RAY DIFFRACTION Resolution:3.85 Å Release Date: 2014-05-28 Classification: RNA |
 |
Speciation Of A Group I Intron Into A Lariat Capping Ribozyme (Heavy Atom Derivative)
Organism: Didymium iridis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-05-28 Classification: RNA Ligands: IRI, MG |
 |
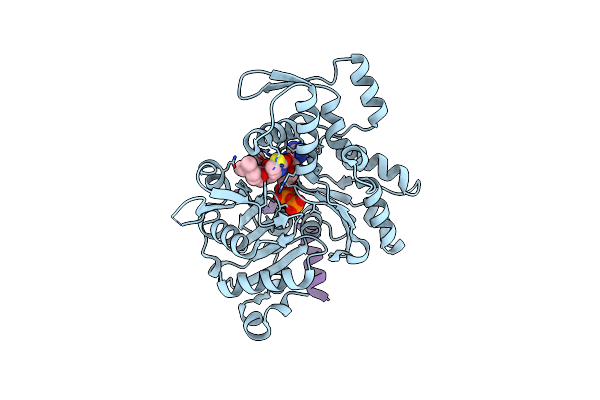
Model Of Full-Length T. Thermophilus Translation Initiation Factor 2 Refined Against Its Cryo-Em Density From A 30S Initiation Complex Map
Organism: Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2013-09-25 Classification: TRANSLATION |
 |
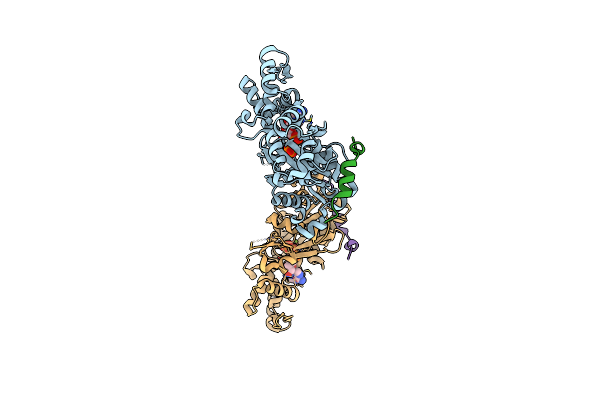
Crystal Structure Of A Chimera Containing The N-Terminal Domain (Residues 8-29) Of Drosophila Ciboulot And The C-Terminal Domain (Residues 18-44) Of Bovine Thymosin-Beta4, Bound To G-Actin-Atp-Latrunculin A
Organism: Drosophila melanogaster, Bos taurus, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-01-25 Classification: CONTRACTILE PROTEIN, PROTEIN BINDING Ligands: ATP, MG, LAR |
 |
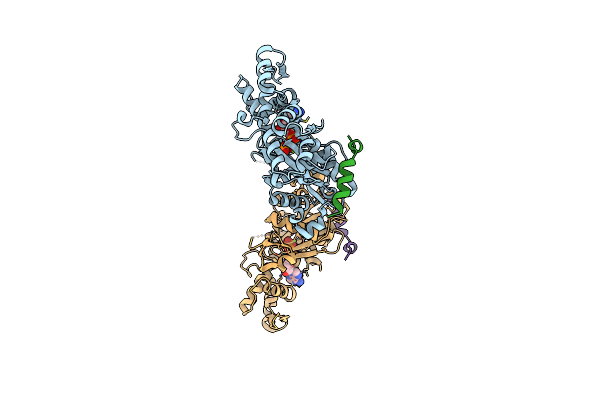
Crystal Structure Of A Chimera Containing The N-Terminal Domain (Residues 8-29) Of Drosophila Ciboulot And The C-Terminal Domain (Residues 18-44) Of Bovine Thymosin-Beta4, Bound To G-Actin-Atp
Organism: Drosophila melanogaster, Bos taurus, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-25 Classification: CONTRACTILE PROTEIN, PROTEIN BINDING Ligands: ATP, MG |
 |
Crystal Structure Of A Chimera Containing The N-Terminal Domain (Residues 8-24) Of Drosophila Ciboulot And The C-Terminal Domain (Residues 13-44) Of Bovine Thymosin-Beta4, Bound To G-Actin-Atp
Organism: Drosophila melanogaster, Bos taurus, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-01-25 Classification: CONTRACTILE PROTEIN, PROTEIN BINDING Ligands: ATP, MG |

