Search Count: 19
 |
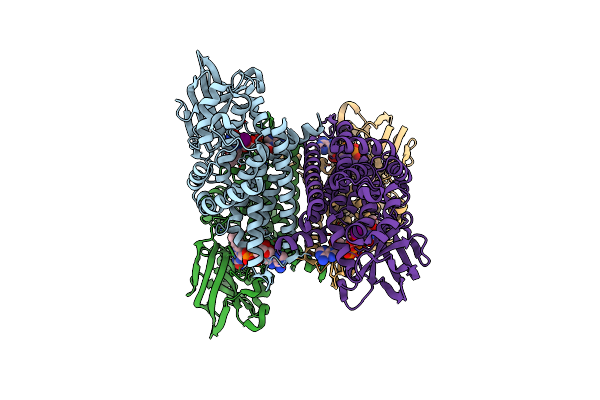
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-05-22 Classification: IMMUNE SYSTEM Ligands: FLC, PO4, 16P, PGE, DXE, FWN, NA |
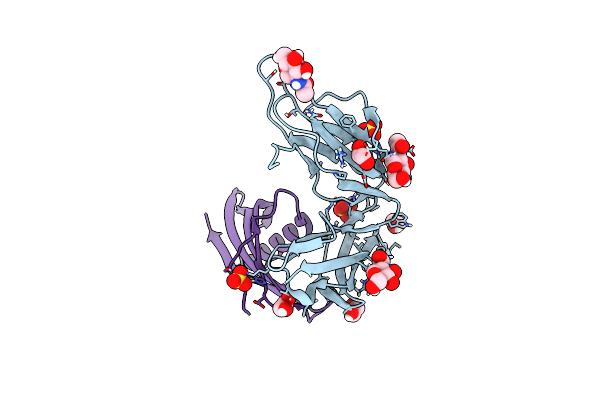 |
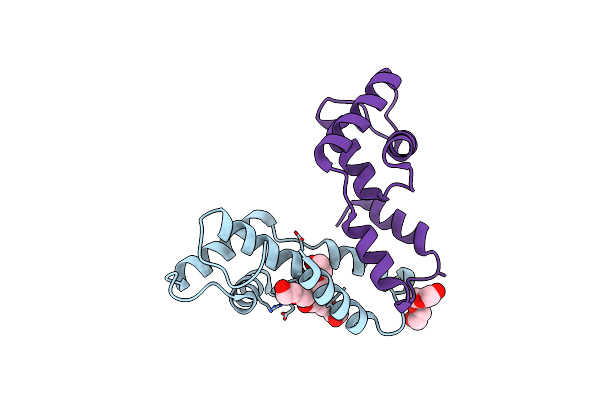
Cocrystal Structure Of Fc Gamma Receptor Iiia Interacting With Affimer F4, A Specific Binding Protein Which Blocks Igg Binding To The Receptor.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-13 Classification: IMMUNE SYSTEM Ligands: NAG, SO4, PEG, CL |
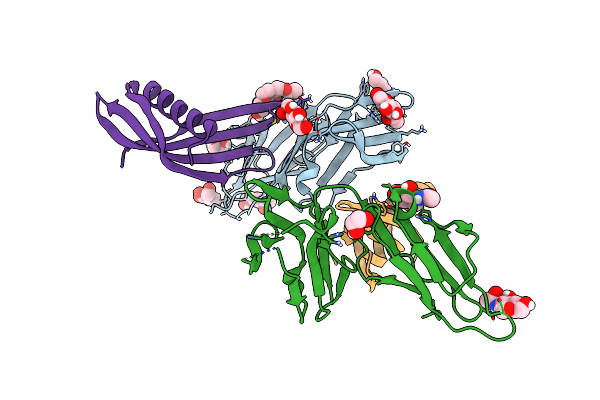 |
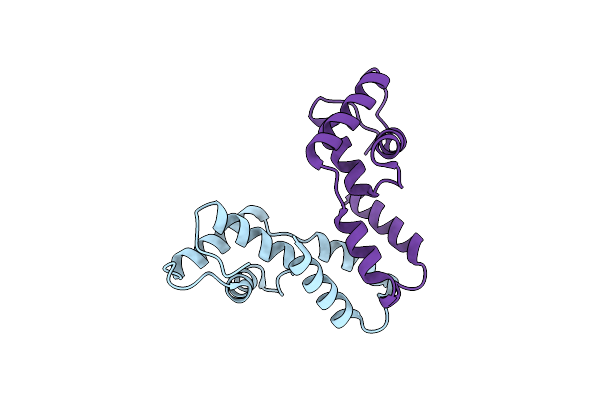
Cocrystal Structure Of Fc Gamma Receptor Iiia Interacting With Affimer G3, A Specific Binding Protein Which Blocks Igg Binding To The Receptor.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-13 Classification: IMMUNE SYSTEM Ligands: NAG, PEG, GOL, PG4 |
 |
Solution Structure Of A Duf3349 Annotated Protein From Mycobacterium Abscessus, Mab_3403C. Seattle Structural Genomics Center For Infectious Disease Target Myaba.17112.A.A2
Organism: Mycobacterium abscessus
Method: SOLUTION NMR Release Date: 2012-11-21 Classification: UNKNOWN FUNCTION |
 |
Solution Structure Of Msmeg_1053, The Second Duf3349 Annotated Protein In The Genome Of Mycobacterium Smegmatis, Seattle Structural Genomics Center For Infectious Disease Target Mysma.17112.B
Organism: Mycobacterium smegmatis str. mc2 155
Method: SOLUTION NMR Release Date: 2011-11-16 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of Polymerase Basic Protein 2 E538-R753 From Influenza A Virus A/Yokohama/2017/03 H3N2
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-03-30 Classification: VIRAL PROTEIN Ligands: IOD, BO3 |
 |
Crystal Structure Of A Putative Fructose-1,6-Biphosphate Aldolase From Coccidioides Immitis Solved By Combined Sad Mr
Organism: Coccidioides immitis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-12-15 Classification: LYASE Ligands: IOD, ZN |
 |
Crystal Structure Of Phosphoserine Phosphatase Serb From Mycobacterium Avium, Native Form
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-11-10 Classification: HYDROLASE Ligands: MG, CL, EDO |
 |
Crystal Structure Of An Acyl-Coa Dehydrogenase From Mycobacterium Thermoresistibile Bound To Reduced Flavin Adenine Dinucleotide Solved By Combined Iodide Ion Sad Mr
Organism: Mycobacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-11-10 Classification: OXIDOREDUCTASE Ligands: FDA, IOD |
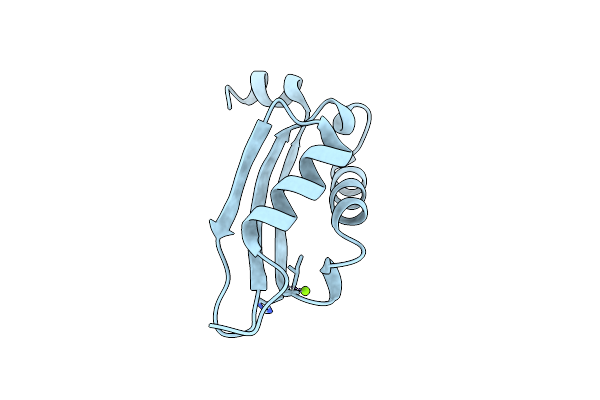 |
Organism: Babesia bovis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-10-13 Classification: UNKNOWN FUNCTION Ligands: IOD, MG |
 |
Crystal Structure Of A Putative Uncharacterized Protein From Mycobacterium Smegamtis, An Ortholog Of Rv0543C, Iodide Phased
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-09-08 Classification: UNKNOWN FUNCTION Ligands: IOD, PG4, PGE |
 |
Crystal Structure Of A Putative Uncharacterized Protein From Mycobacterium Smegmatis, An Ortholog Of Rv0543C
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-09-08 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of A Putative Acyl-Coa Dehydrogenase From Mycobacterium Smegmatis, Iodide Soak
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-09-01 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Enoyl-Coa Hydratase From Mycobacterium Smegmatis, Iodide Soak
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-07-28 Classification: LYASE Ligands: IOD |
 |
Crystal Structure Of Acetylpolyamine Aminohydrolase From Burkholderia Pseudomallei, Iodide Soak
Organism: Burkholderia pseudomallei 1710b
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: ZN, SO4, K, IOD |
 |
Crystal Structure Of Extragenic Suppressor Protein Suhb From Bartonella Henselae, Via Combined Iodide Sad Molecular Replacement
Organism: Bartonella henselae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: MG, IOD |
 |
Crystal Structure Of Alanine Racemase From Bartonella Henselae With Covalently Bound Pyridoxal Phosphate
Organism: Bartonella henselae
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2009-12-22 Classification: ISOMERASE |
 |
Crystal Structure Of Eoxycytidine Triphosphate Deaminase From Anaplasma Phagocytophilum At 2.1A Resolution
Organism: Anaplasma phagocytophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-24 Classification: HYDROLASE Ligands: IOD |
 |
Crystal Structure Of A Plasmid Partition Protein From Borrelia Burgdorferi At 2.25A Resolution, Iodide Soak
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-11-03 Classification: BIOSYNTHETIC PROTEIN |

