Search Count: 140
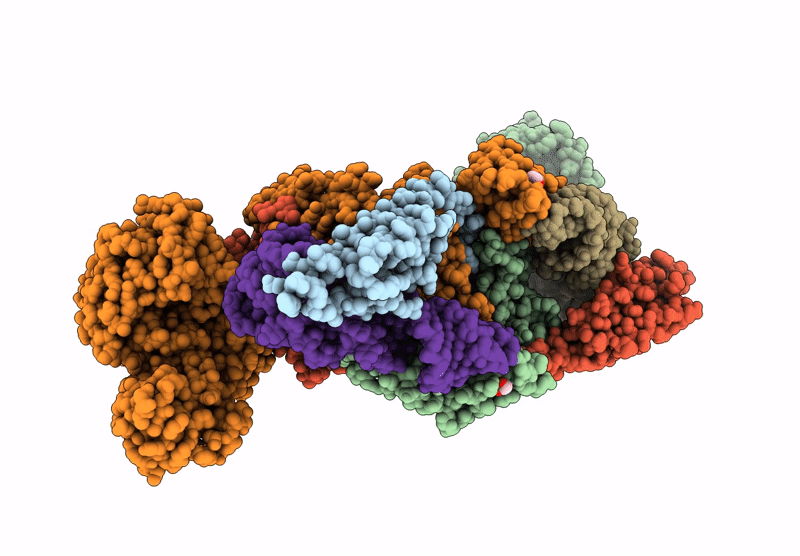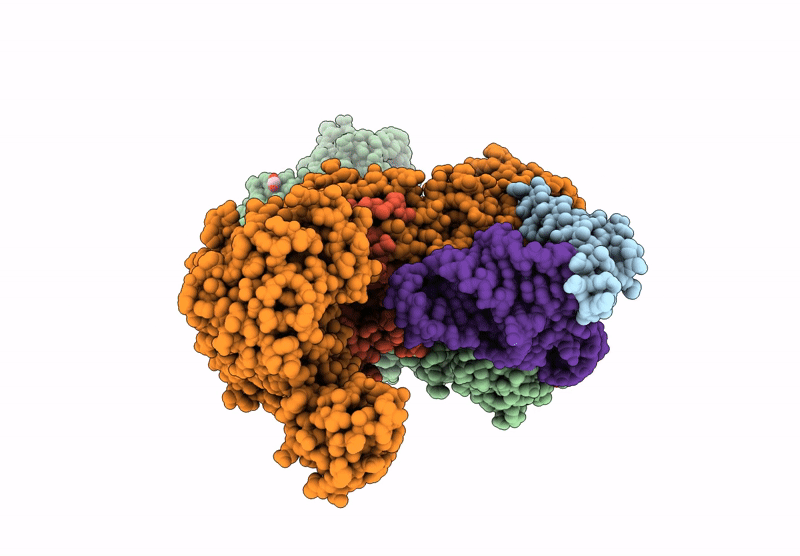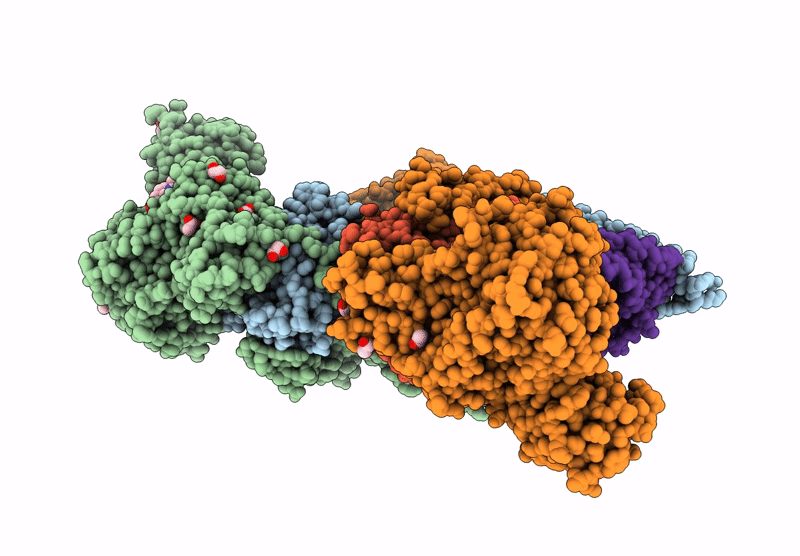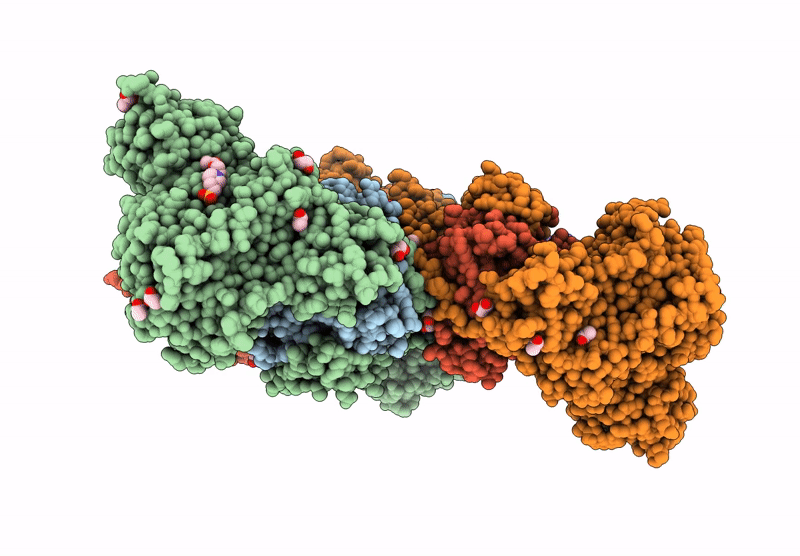 |
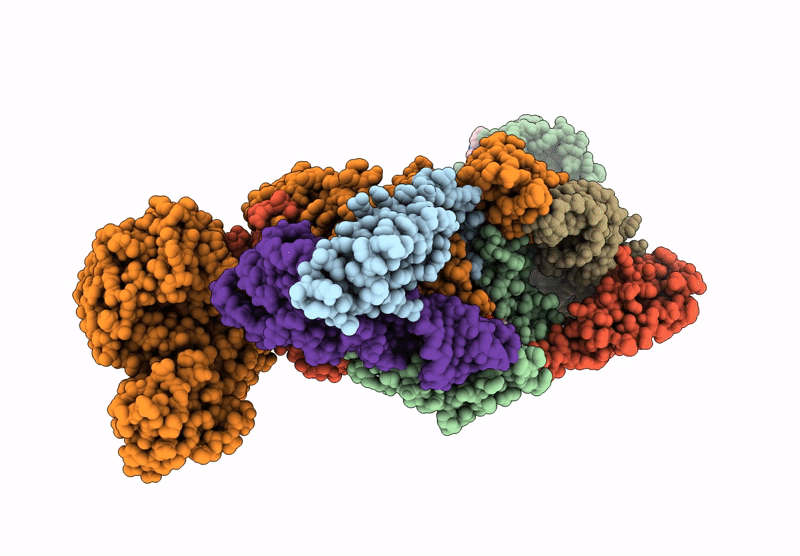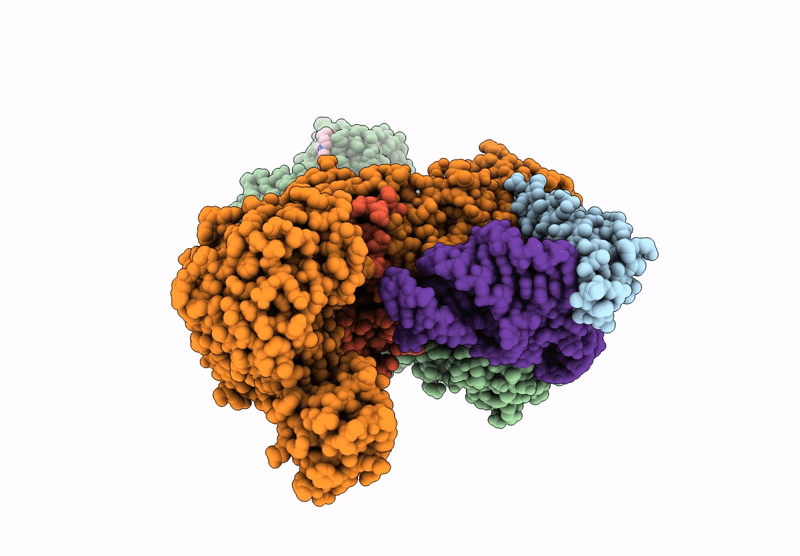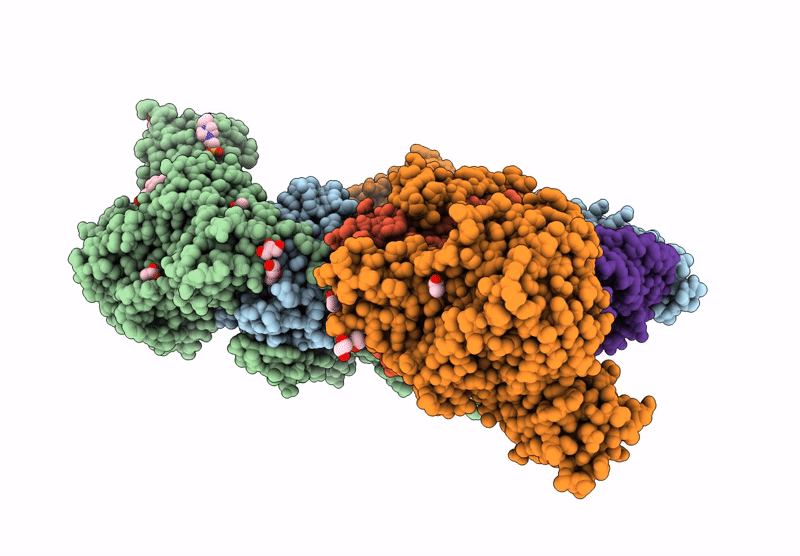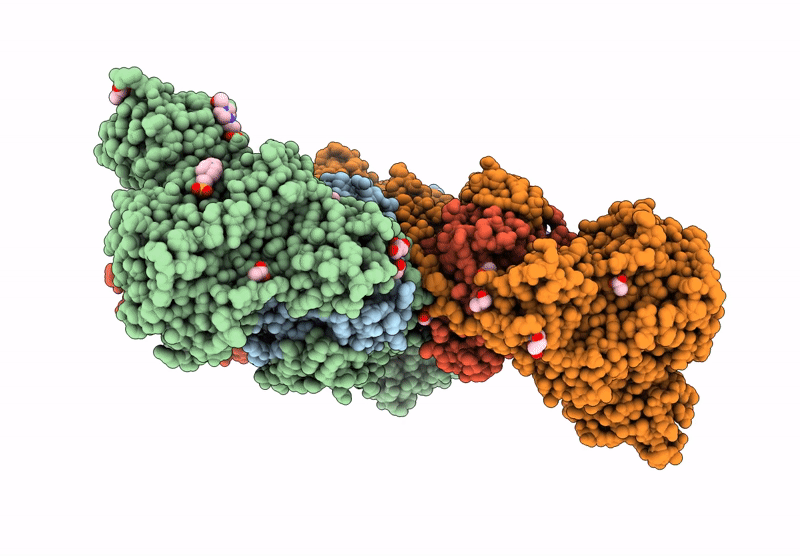
Crystal Structure Of M. Tuberculosis Phers-Trna Complex Bound To Inhibitor D-116
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-02-12 Classification: LIGASE/RNA Ligands: AX7, MG, ACT, EDO, PEG, EPE, DMS |
 |
Crystal Structure Of M. Tuberculosis Phers-Trna Complex Bound To Inhibitor D-004
Organism: Mycobacterium tuberculosis h37rv, Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-02-12 Classification: LIGASE Ligands: 2AQ, MG, ACT, PEG, GOL, DMS, EPE, EDO |
 |
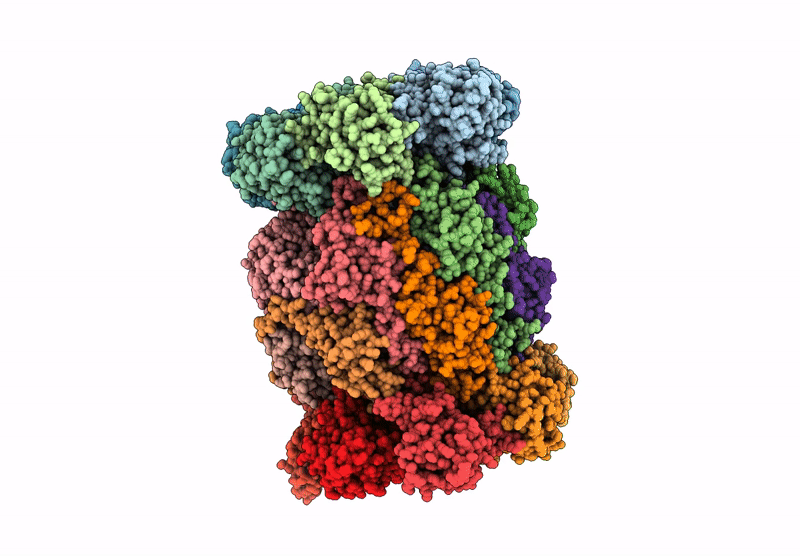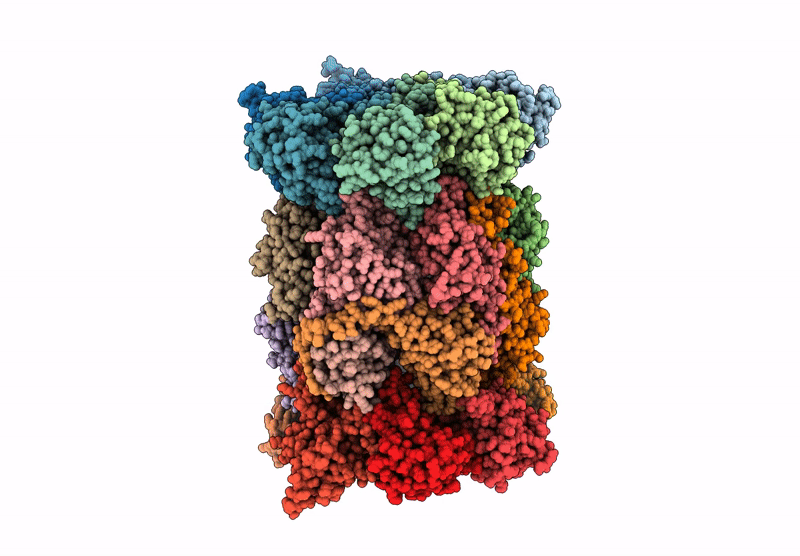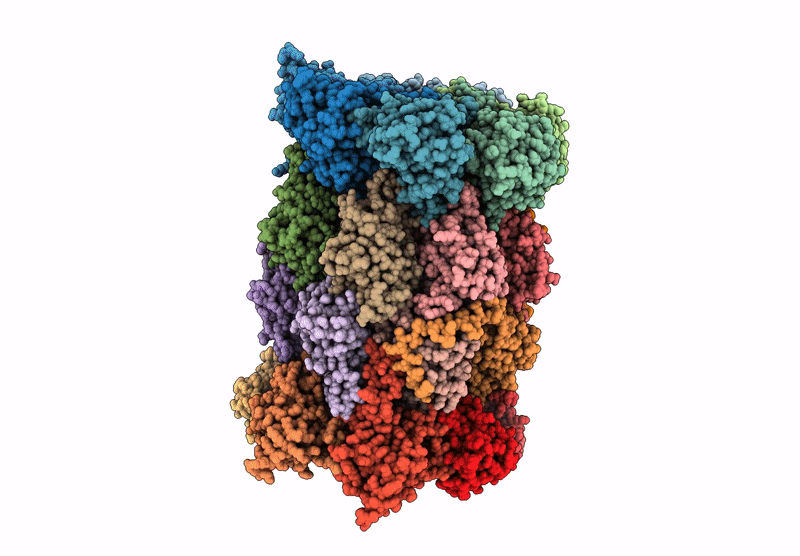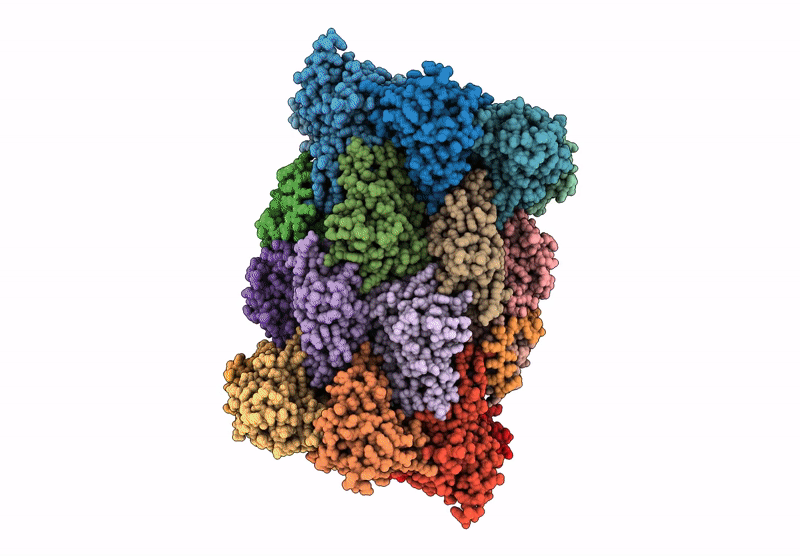
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: ELECTRON TRANSPORT, Translocase Ligands: MG, DGT, NDP, ZN, EHZ, HEM, HEC, FMN, SF4, FES |
 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2024-12-25 Classification: UNKNOWN FUNCTION |
 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Resolution:2.31 Å Release Date: 2024-12-25 Classification: UNKNOWN FUNCTION |
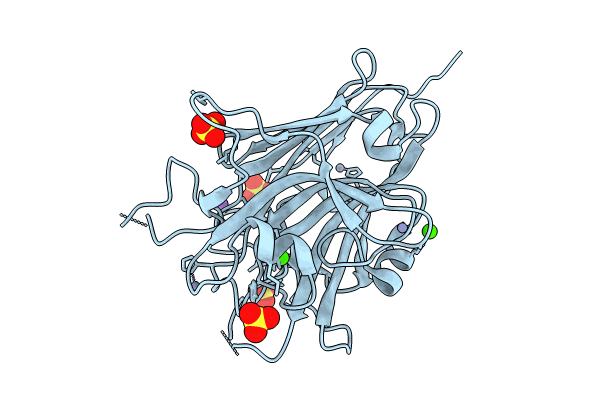 |
Organism: Staphylococcus aureus subsp. aureus mw2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-05-08 Classification: CELL ADHESION Ligands: SO4, CA, NA, ZN |
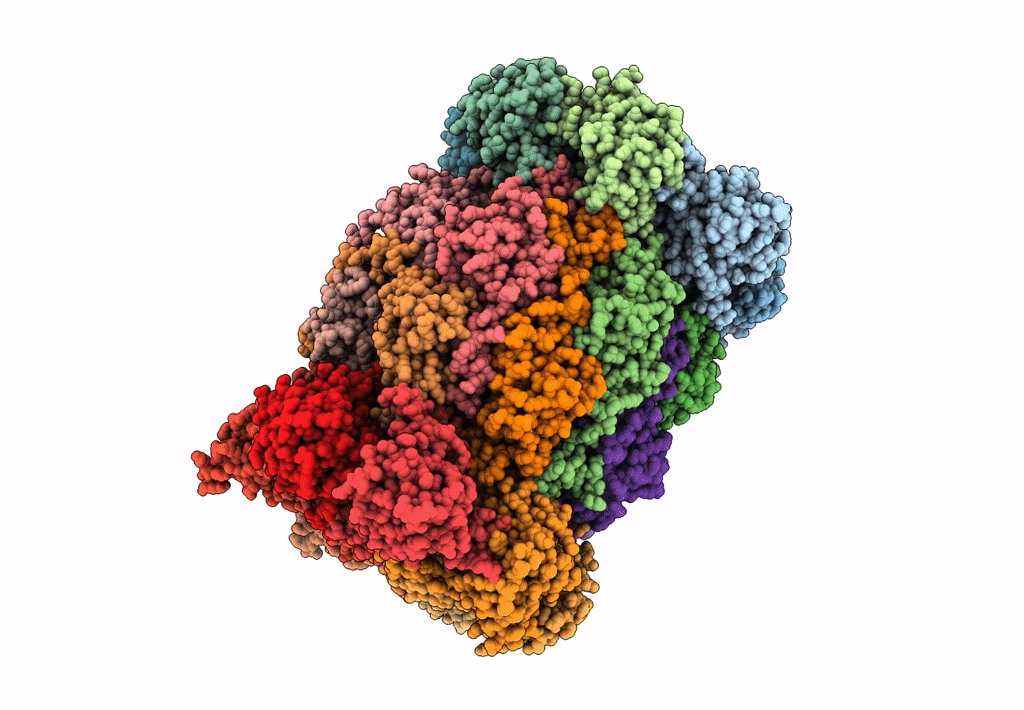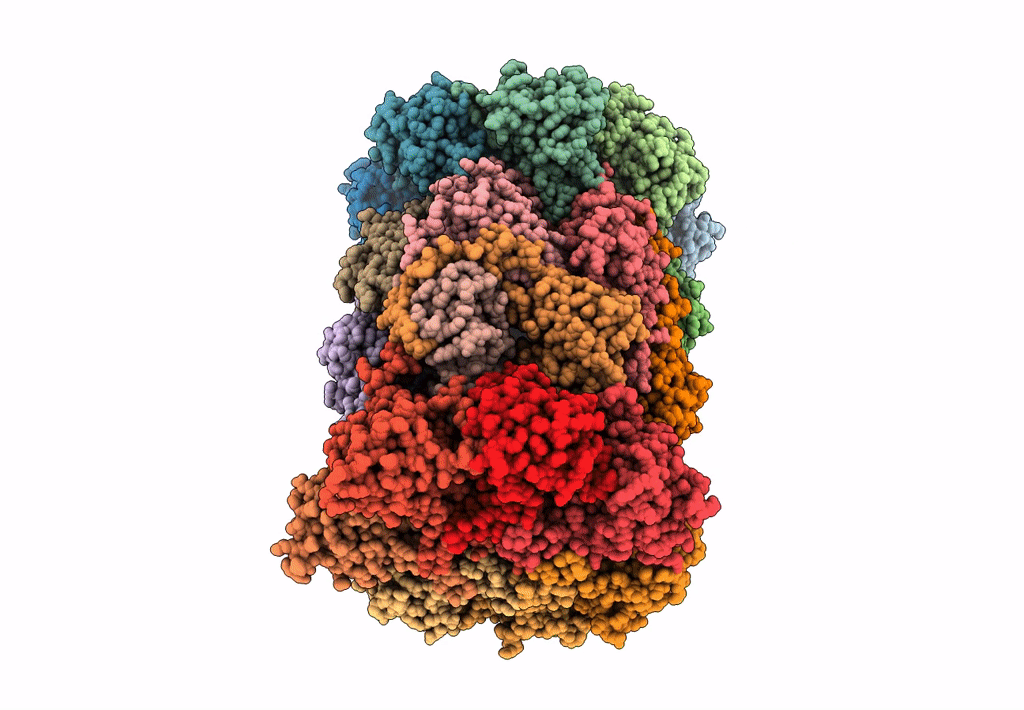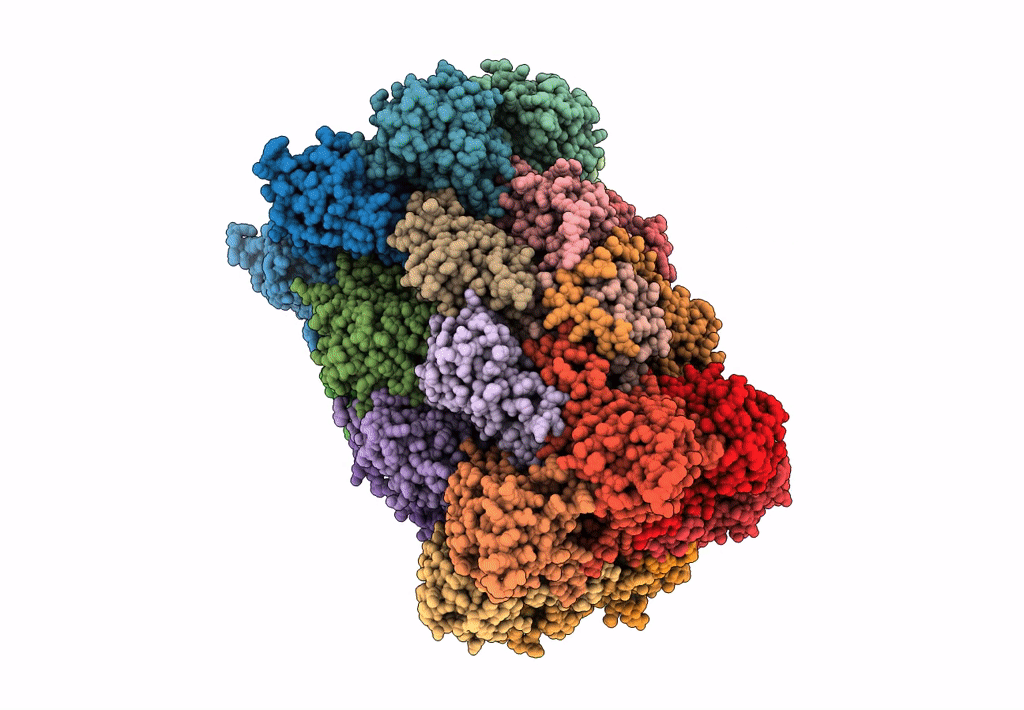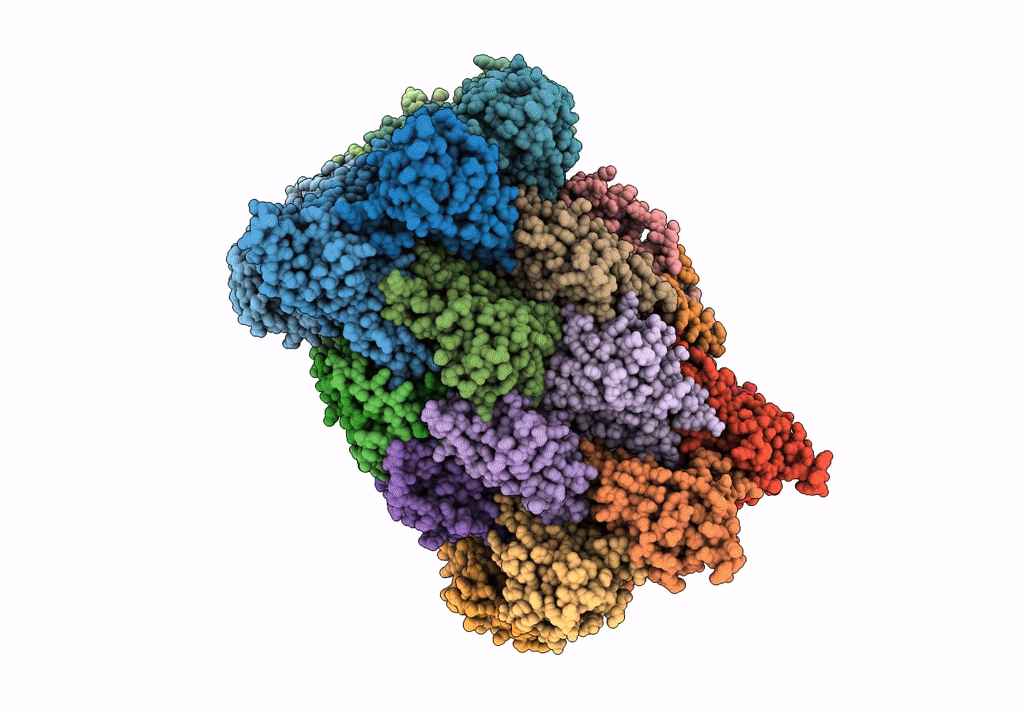 |
Leishmania Tarentolae Proteasome 20S Subunit In Complex With 1-Benzyl-N-(3-(Cyclopropylcarbamoyl)Phenyl)-6-Oxo-1,6-Dihydropyridazine-3-Carboxamide
Organism: Leishmania tarentolae
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2023-08-09 Classification: UNKNOWN FUNCTION Ligands: VYW |
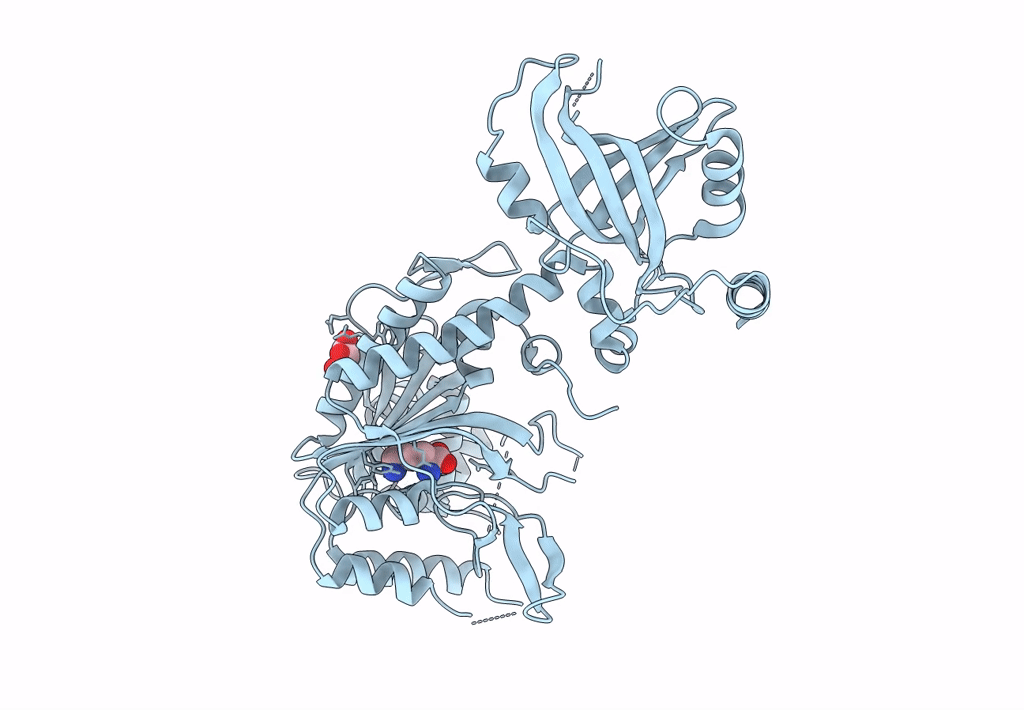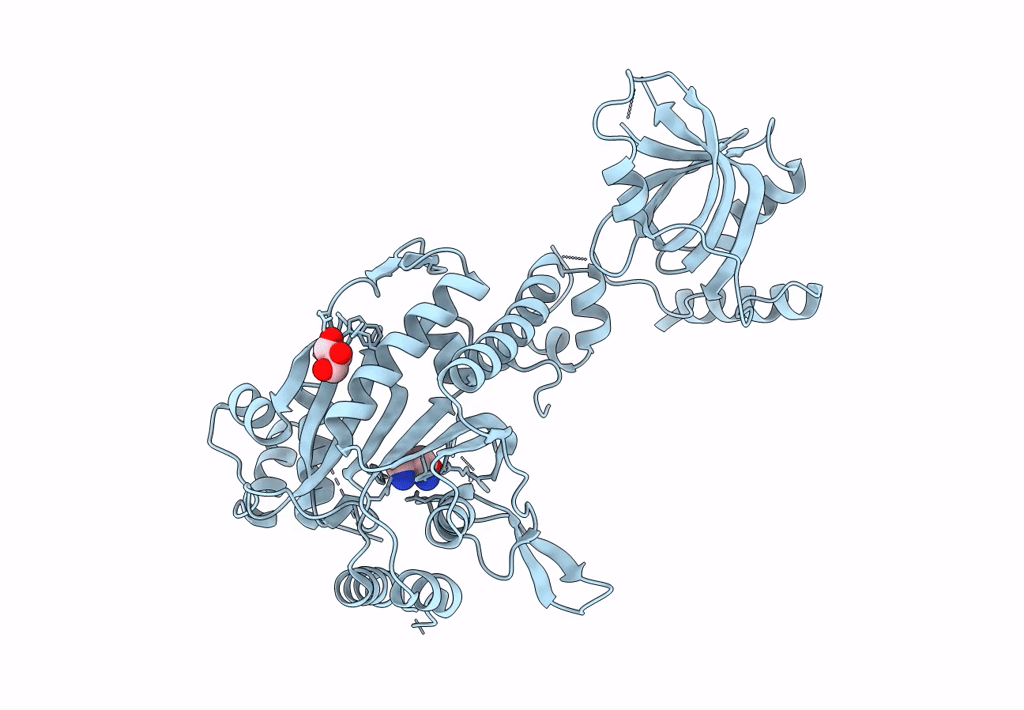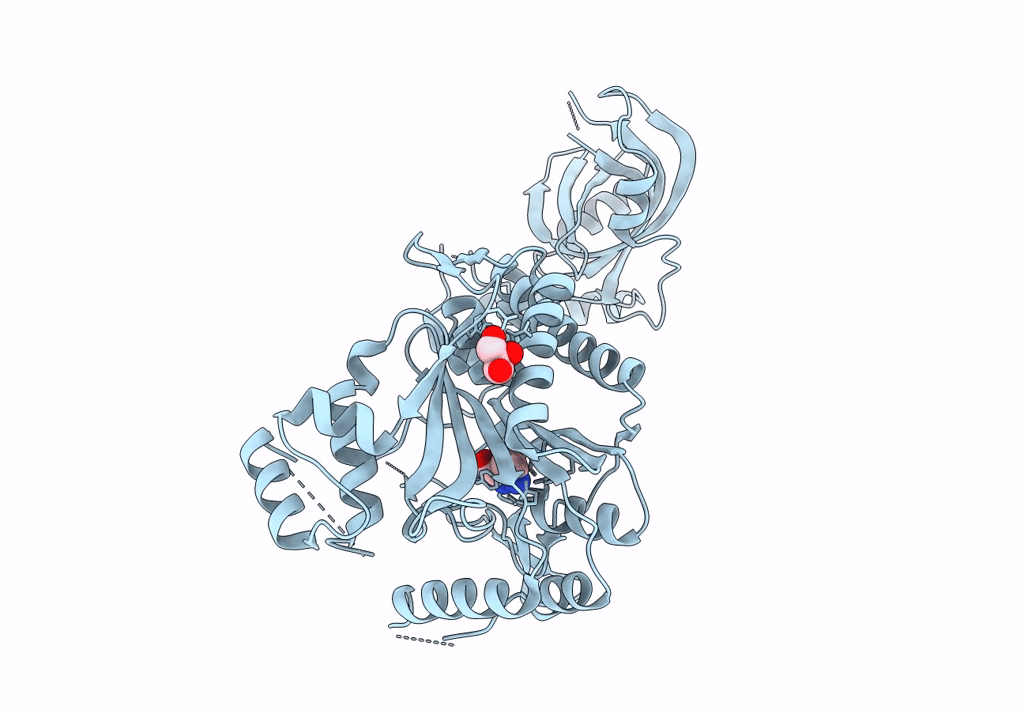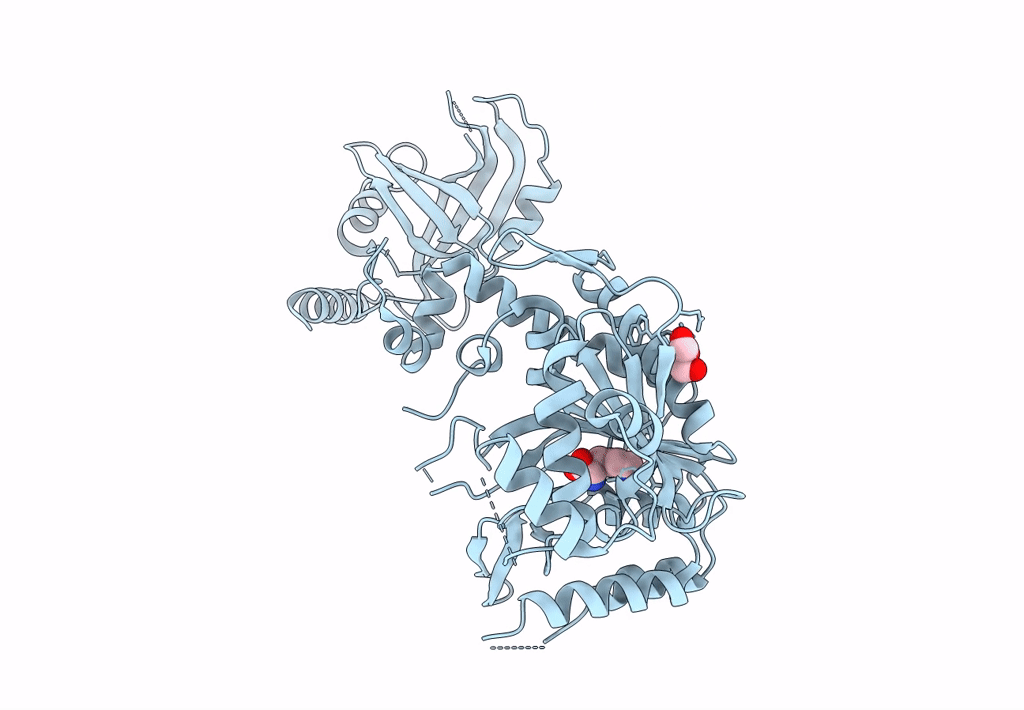 |
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, GOL |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine And An Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, C6I |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine And Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, DD0 |
 |
Crystal Structure Of Quinonoid Dihydropteridine Reductase From Leishmania Donovani
Organism: Leishmania donovani
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE |
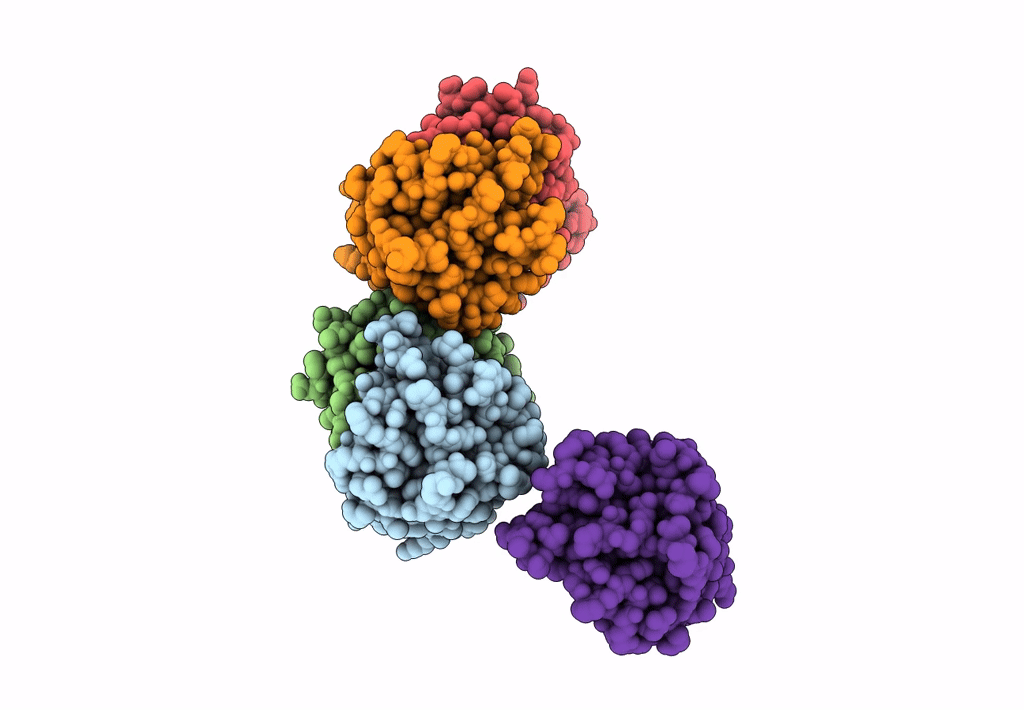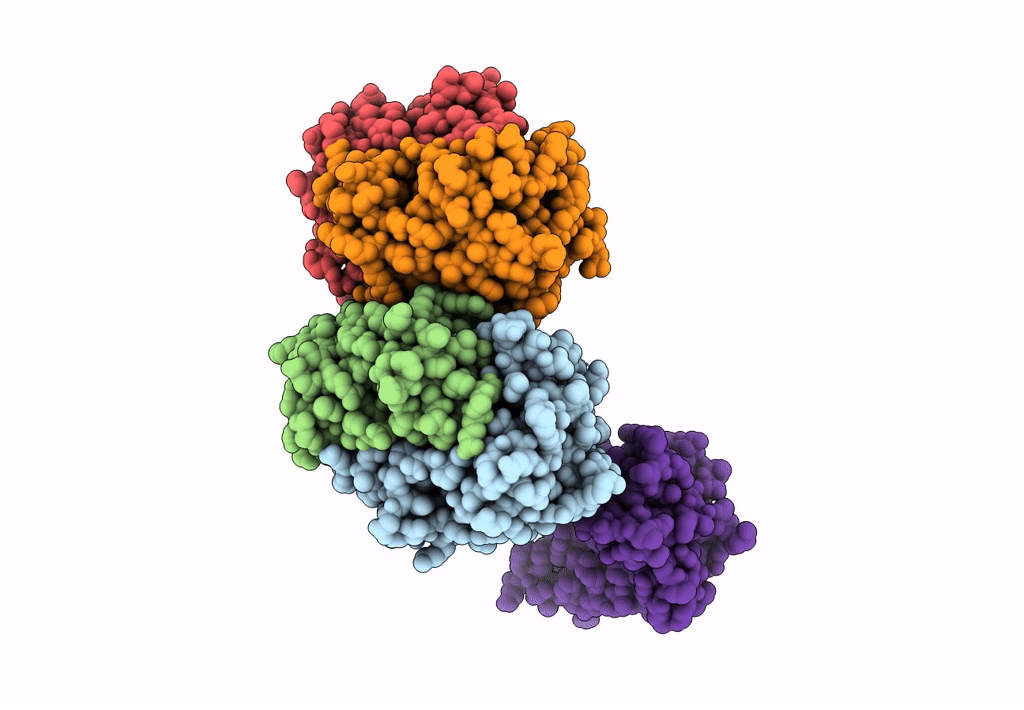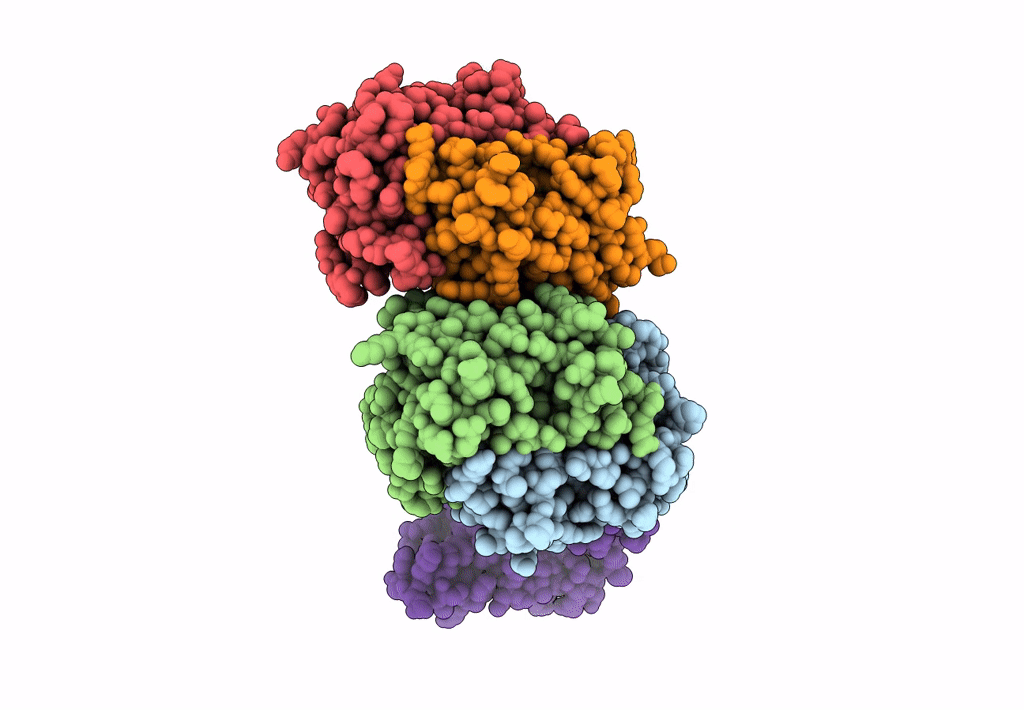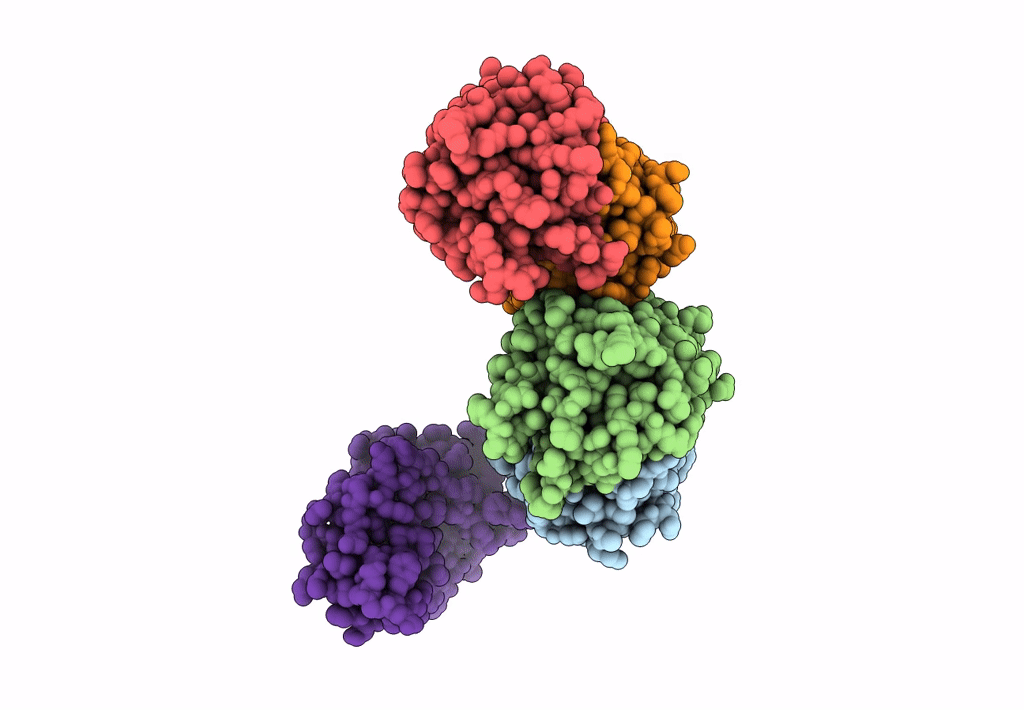 |
Crystal Structure Of 3-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase (Faba) From Pseudomonas Aeruginosa In Complex With Z30857828
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-10-19 Classification: LYASE Ligands: M25, DMS |
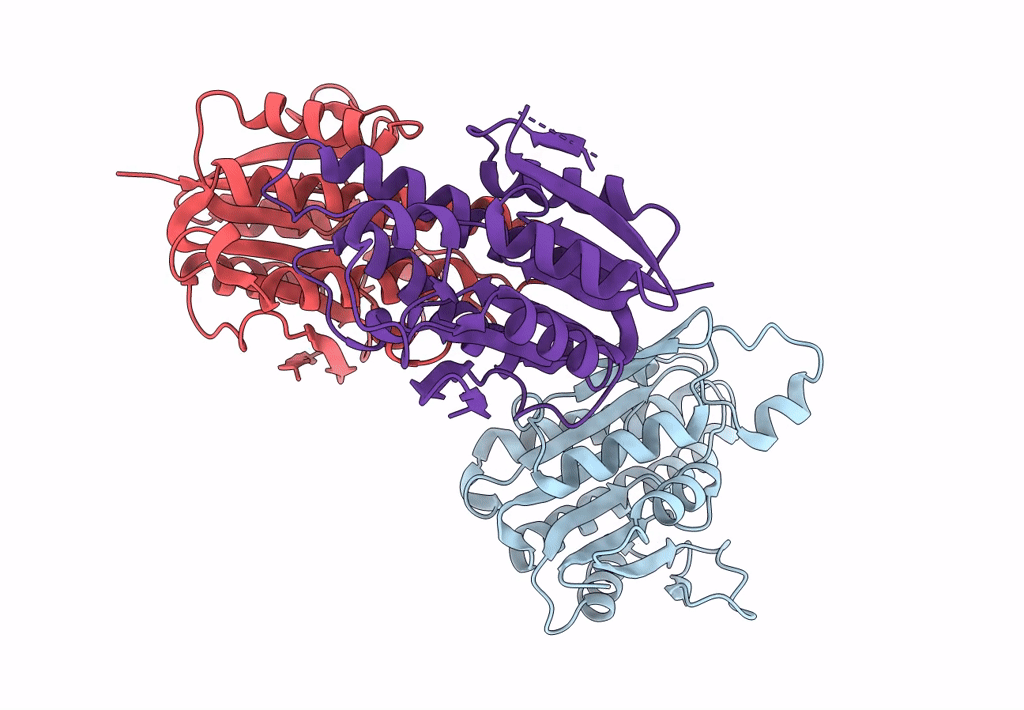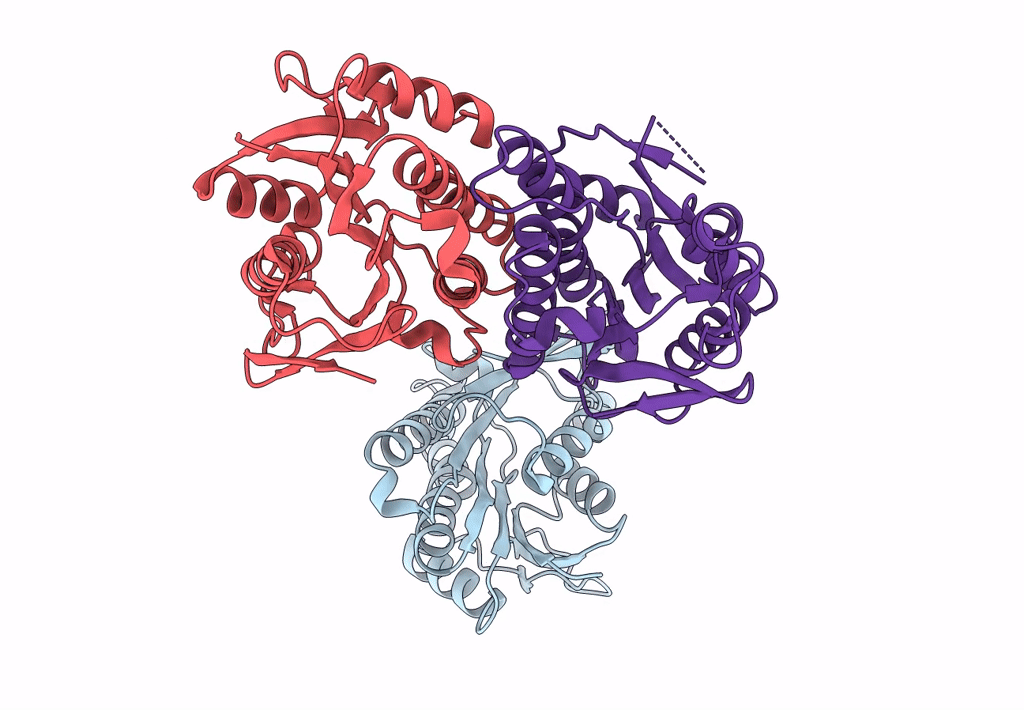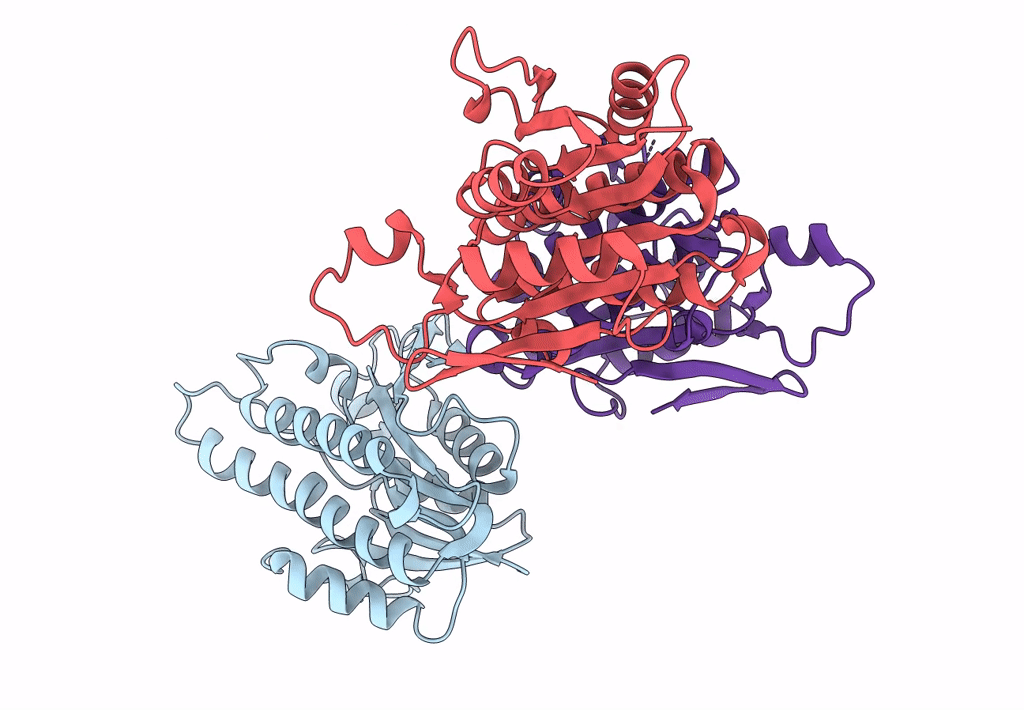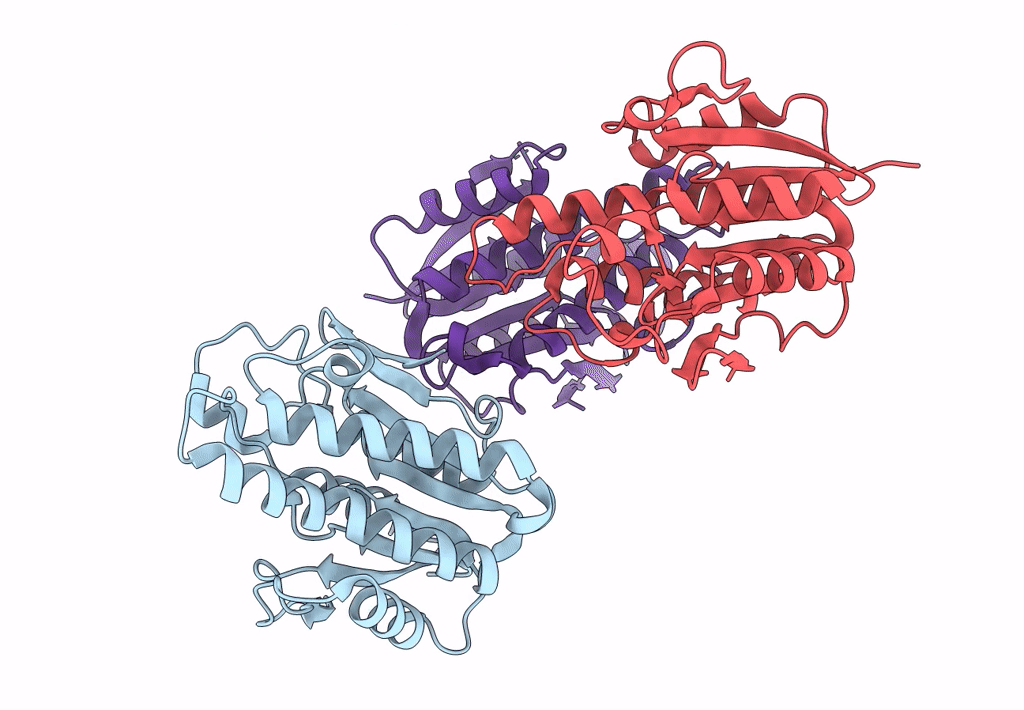 |
Crystal Structure Of Quinonoid Dihydropteridine Reductase From Leishmania Major
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-05 Classification: OXIDOREDUCTASE |
 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-10-05 Classification: ISOMERASE Ligands: MG, PO4 |
 |
Crystal Structure Of Cryptosporidium Parvum -Plasmodium Falciparum Mutant Lysyl Trna Synthetase In Complex With Inhibitor
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: LIGASE Ligands: LYS, JE5, GOL |
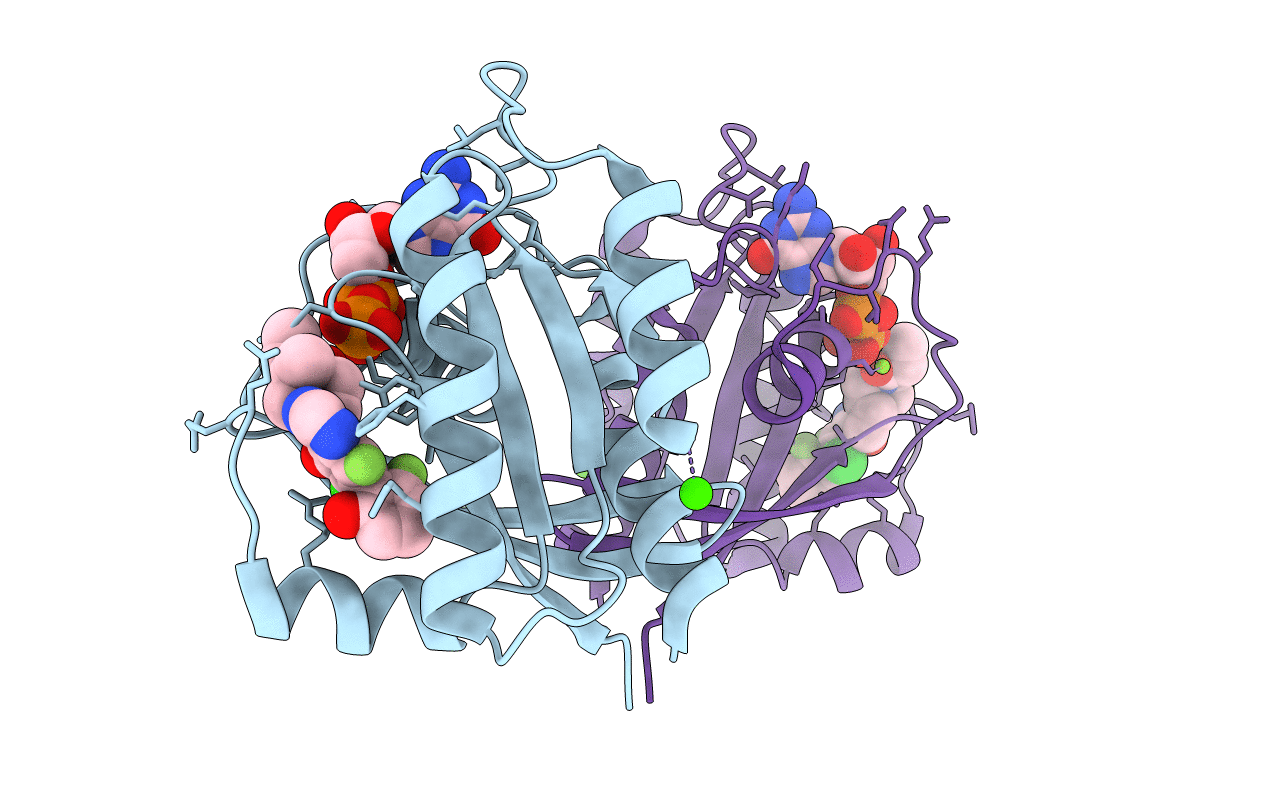 |
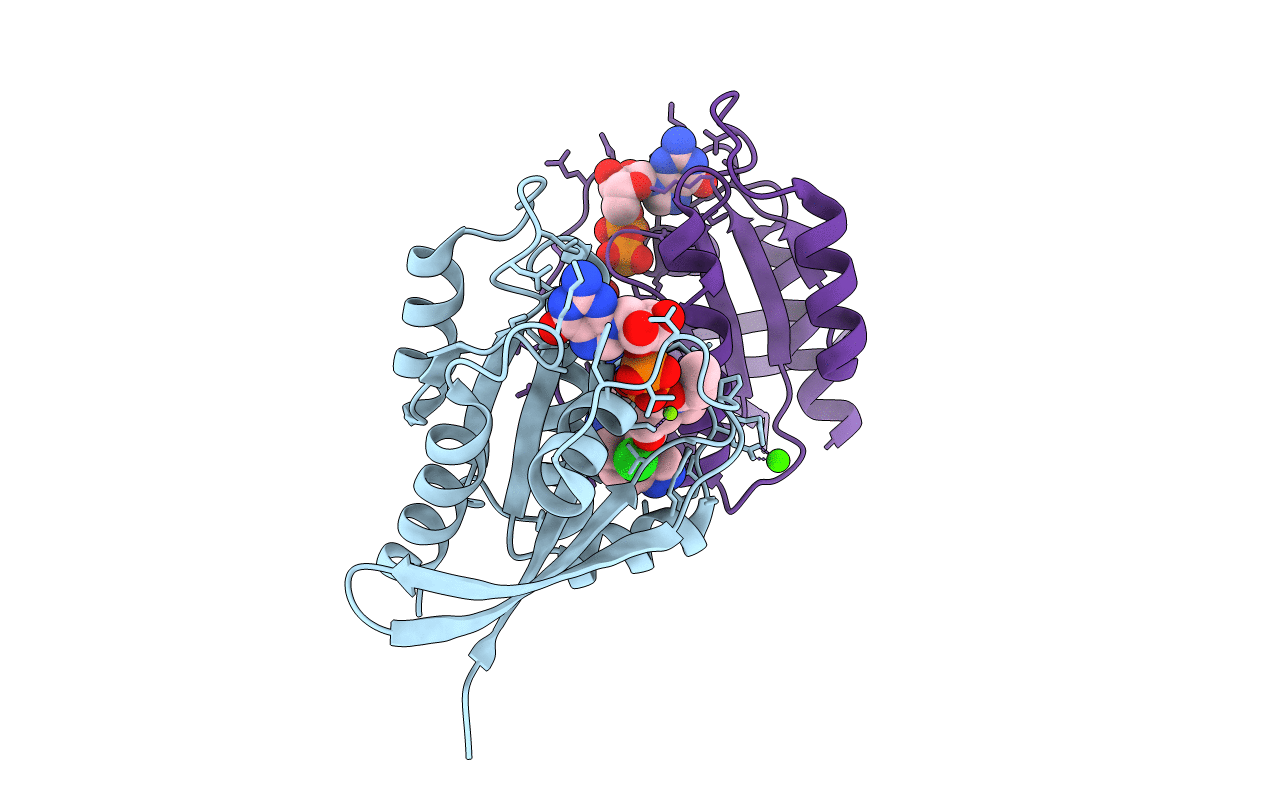
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2022-04-20 Classification: SIGNALING PROTEIN Ligands: V4T, MG, GDP, CA |
 |
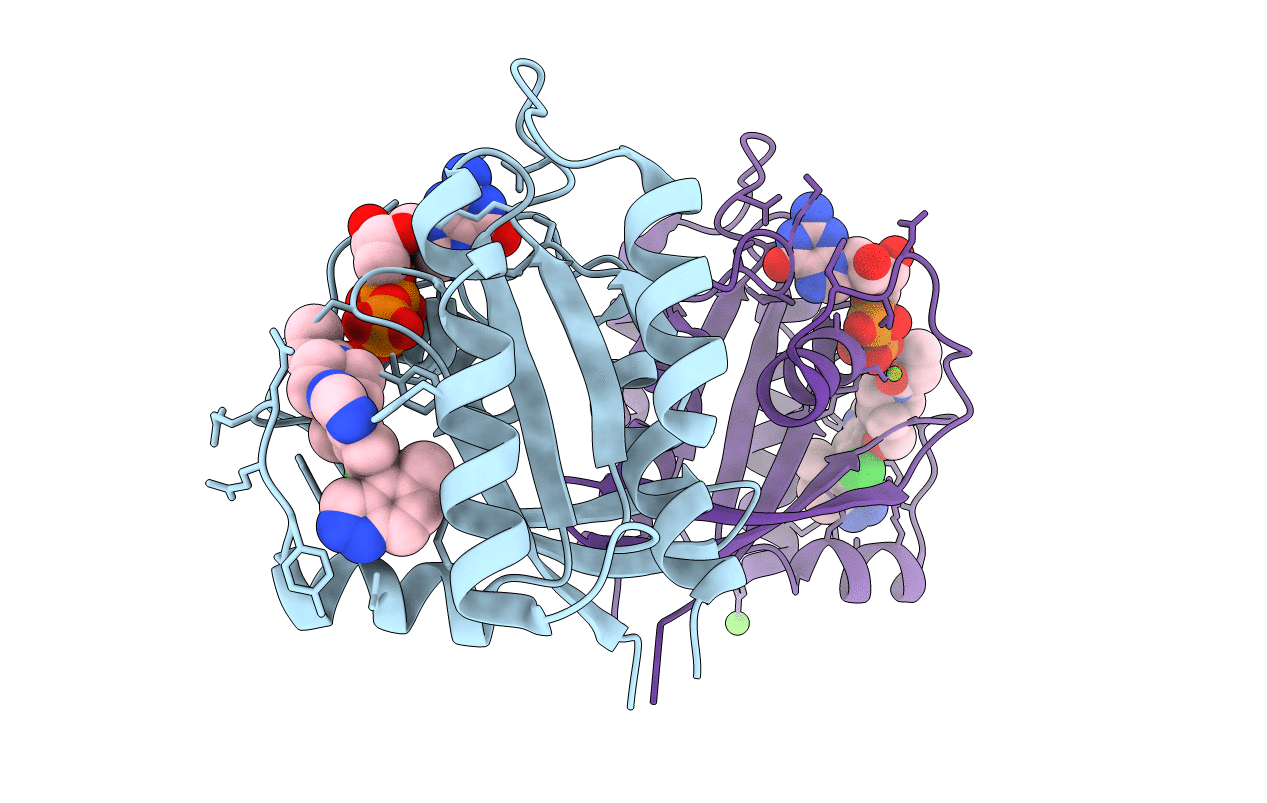
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-04-20 Classification: SIGNALING PROTEIN Ligands: MG, GDP, V52, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-04-20 Classification: SIGNALING PROTEIN Ligands: MG, GDP, VLE, CA |
 |
Crystal Structure Of 3-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase (Faba) From Pseudomonas Aeruginosa In Complex With Ddd00084774
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-03-02 Classification: LYASE Ligands: TQH |
 |
Crystal Structure Of 3-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase (Faba)From Pseudomonas Aeruginosa In Complex With Ddd00082063
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-03-02 Classification: LYASE Ligands: TZQ |

