Search Count: 131
 |
The Inhibitor Of Toll-Like Receptor Signaling O-Vanillin Binds Covalently To Mal/Tirap Lys-210
|
 |
 |
Co-Crystal Structure Of The Sars-Cov2 Main Protease Nsp5 With An Uracil-Carrying X77-Like Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-01-31 Classification: ANTIVIRAL PROTEIN Ligands: MLI, YQN, NA, DMS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: GENE REGULATION Ligands: ZN |
 |
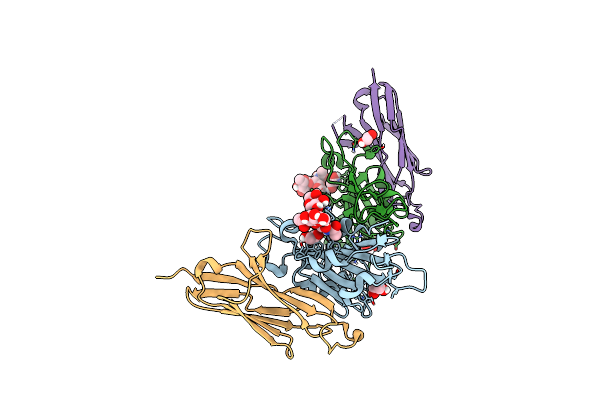
Aicar Transformylase/Imp Cyclohydrolase (Atic) Is Essential For De Novo Purine Biosynthesis And Infection By Cryptococcus Neoformans
Organism: Cryptococcus neoformans
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-04-20 Classification: STRUCTURAL PROTEIN Ligands: MG |
 |
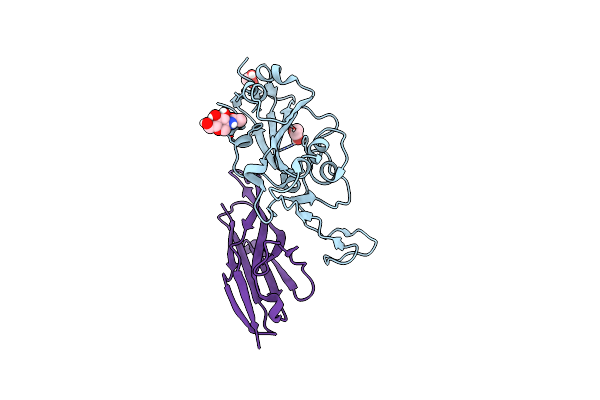
Crystal Structure Of The Sars-Cov-2 Receptor Binding Domain In Complex With Vnar 3B4
Organism: Severe acute respiratory syndrome coronavirus 2, Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-01-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: EDO |
 |
Crystal Structure Of The Sars-Cov-2 Receptor Binding Domain In Complex With Vnar 2C02
Organism: Severe acute respiratory syndrome coronavirus 2, Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-01-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: EDO, NAG, CL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FXU, ZN, MG, CL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FXX, MPO, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FY6, ZN, CL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FY9, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYF, ZN, MG |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYL, PO4, EDO, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: MPB, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FYR, DMS, ZN |
 |
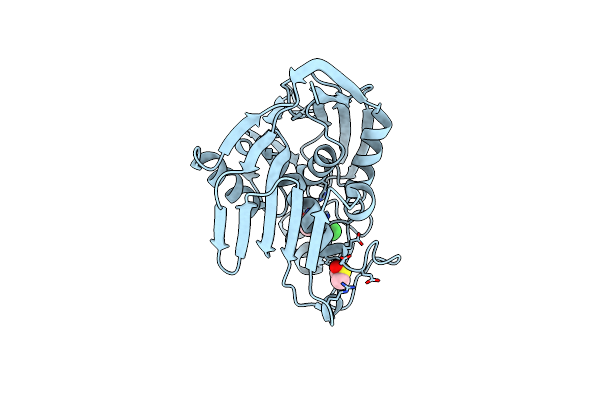
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ0, ZN |
 |
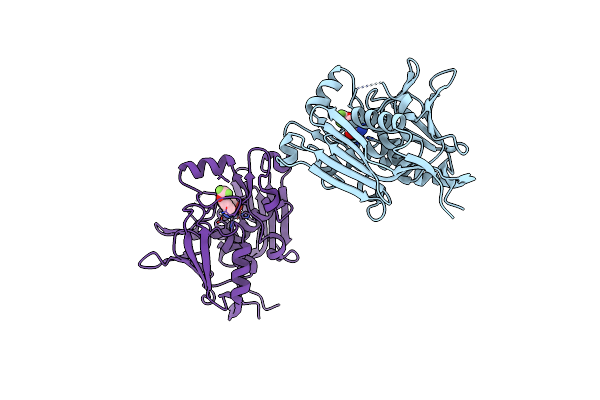
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ3, DMS, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: FZ6, ZN |
 |
P146A Variant Of Beta-Phosphoglucomutase From Lactococcus Lactis In Complex With Glucose 6-Phosphate And Trifluoromagnesate
Organism: Lactococcus lactis subsp. lactis (strain il1403)
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2020-10-28 Classification: ISOMERASE Ligands: BG6, MGF, MG, NA, EDO, PDO |
 |
Substrate-Free P146A Variant Of Beta-Phosphoglucomutase From Lactococcus Lactis
Organism: Lactococcus lactis subsp. lactis (strain il1403)
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2020-10-28 Classification: ISOMERASE Ligands: MG |

