Search Count: 40
 |
Crystal Structure Of A Pentafluoro-Phe Incorporated Phosphatidylinositol-Specific Phospholipase C (H258X)From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-01 Classification: LYASE Ligands: ACT, INS |
 |
Structure Of The F249X Mutant Of Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus
Organism: Staphylococcus aureus str. newman
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-07-01 Classification: LYASE Ligands: INS, ACT |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-18 Classification: TRANSFERASE Ligands: 1KH, CL, NA, PO4, GOL |
 |
Structure Of The Unliganded N254Y/H258Y Mutant Of The Phosphatidylinositol-Specific Phospholipase C From S. Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-04-10 Classification: hydrolase, lyase Ligands: CL, ACT |
 |
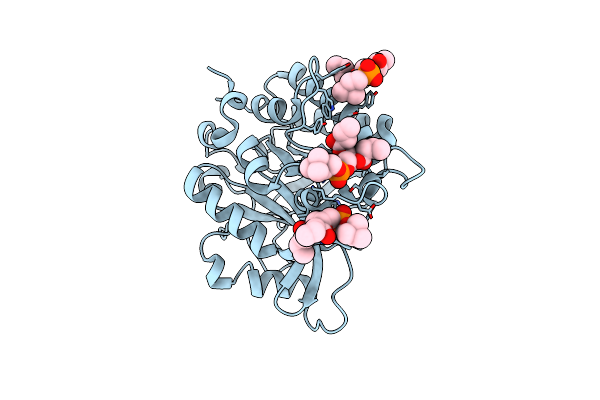
Structure Of The N254Y/H258Y Mutant Of The Phosphatidylinositol-Specific Phospholipase C From S. Aureus Bound To Choline
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-04-10 Classification: hydrolase, lyase Ligands: ACT, CHT, CL |
 |
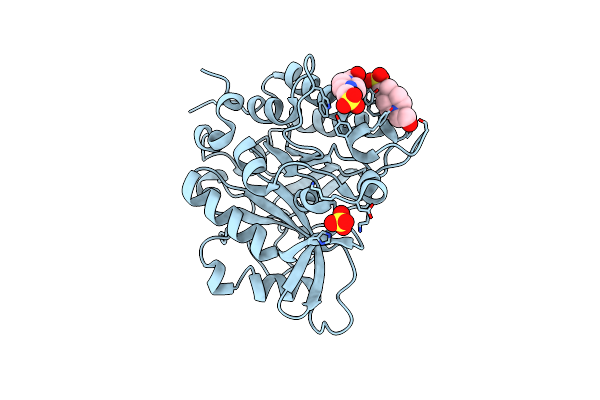
Structure Of The N254Y/H258Y Mutant Of The Phosphatidylinositol-Specific Phospholipase C From S. Aureus Bound To Dic4Pc
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-04-10 Classification: hydrolase, lyase Ligands: ACT, XP5 |
 |
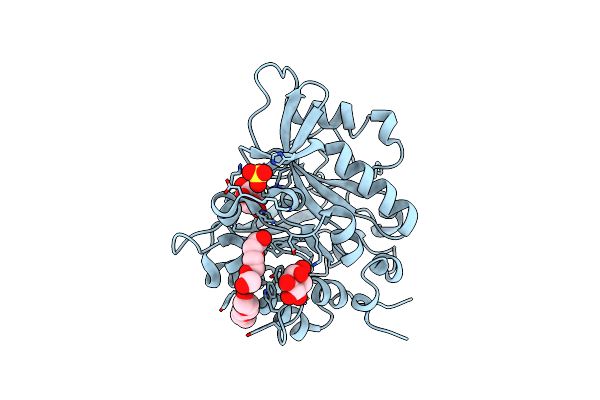
Structure Of The N254Y/H258Y Mutant Of The Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus Bound To Hepes
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-10 Classification: hydrolase, lyase Ligands: EPE, SO4 |
 |
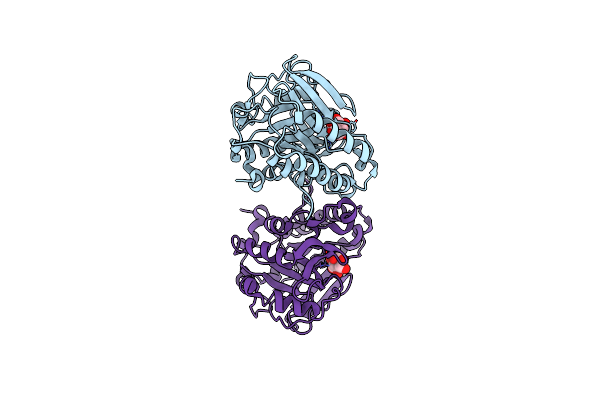
Structure Of The H258Y Mutant Of The Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-04-10 Classification: hydrolase, lyase Ligands: SO4, INS, PGE |
 |
Modulation Of S.Aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2012-12-12 Classification: LYASE Ligands: INS |
 |
Modulation Of S.Aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding.
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-12-12 Classification: LYASE Ligands: ACT |
 |
Structure Of The N254Y/H258Y Double Mutant Of The Phosphatidylinositol-Specific Phospholipase C From S.Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-12-12 Classification: LYASE Ligands: EPE, SO4 |
 |
An Intramolecular Pi-Cation Latch In Phosphatidylinositol-Specific Phospholipase C From S.Aureus Controls Substrate Access To The Active Site
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-04-04 Classification: LYASE Ligands: INS, CL |
 |
Structure Of The Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2012-04-04 Classification: LYASE Ligands: SO4, IPA |
 |
Structure Of The H258Y Mutant Of Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-04-04 Classification: LYASE Ligands: INS, ACT |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-01-11 Classification: ISOMERASE Ligands: PO4, NAD, PG4, GOL, NA |
 |
L-Myo-Inositol 1-Phosphate Synthase From Archaeoglobus Fulgidus Wild-Type With The Intermediate 5-Keto 1-Phospho Glucose
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-11 Classification: ISOMERASE Ligands: PO4, NAI, PG4, KPG, GOL, NA |
 |
L-Myo-Inositol 1-Phosphate Synthase From Archaeoglobus Fulgidus Mutant K278A
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-11 Classification: ISOMERASE Ligands: PO4, NAD, PG4, GOL, NA |
 |
L-Myo-Inositol 1-Phosphate Synthase From Archaeoglobus Fulgidus Mutant K367A
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-11 Classification: ISOMERASE Ligands: PO4, SO4, NAD, GOL, PG4, NA |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2012-01-11 Classification: ISOMERASE Ligands: PO4, NAD, GOL, SO4, K |
 |
Crystal Structure Of The Y247S/Y251S Mutant Of Phosphatidylinositol-Specific Phospholipase C From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-04-14 Classification: LYASE Ligands: ZN |

