Search Count: 40
 |
Structure Of The Pl6 Family Alginate Lyase Patl3640 From Pseudoalteromonas Atlantica T6C
Organism: Pseudoalteromonas atlantica (strain t6c / atcc baa-1087)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2021-07-28 Classification: LYASE Ligands: GOL |
 |
Structure Of The Pl6 Family Chondroitinase B From Pseudopedobacter Saltans, Pedsa3807
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-07-28 Classification: LYASE |
 |
Structure Of The Pl6 Family Polysaccharide Lyase Pedsa3628 From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-07-28 Classification: LYASE Ligands: PO4 |
 |
Structure Of The Pl6 Family Alginate Lyase Pedsa0632 From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-07-28 Classification: LYASE |
 |
Structure Of The Pl6 Family Alginate Lyase Patl3640 From Pseudoalteromonas Atlantica T6C In Complex With 4-Deoxy-L-Erythro-5-Hexoseulose Uronic Acid
Organism: Pseudoalteromonas atlantica (strain t6c / atcc baa-1087)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-07-28 Classification: LYASE Ligands: V4W, GOL, EDO |
 |
Structure Of The Pl6 Family Alginate Lyase Pedsa0632 From Pseudopedobacter Saltans In Complex With Substrate
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-07-28 Classification: LYASE |
 |
Organism: Fasciola hepatica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-12 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-25 Classification: ISOMERASE Ligands: CA, GLC, GOL |
 |
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-09-25 Classification: ISOMERASE Ligands: GOL, CA, SO4 |
 |
Mutb Inactive Double Mutant D200A-D415N Soaked With Sucrose And Having As Bound Ligands Sucrose In Molecule A And The Reaction Product Trehalulose In Molecule B
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-09-25 Classification: ISOMERASE Ligands: GOL, CA, TEU |
 |
Crystal Structure Of The Trehalulose Synthase Mutb In Complex With Trehalulose
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-09-25 Classification: ISOMERASE Ligands: CA, TEU |
 |
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-09-25 Classification: ISOMERASE Ligands: CA, ISL, GLC |
 |
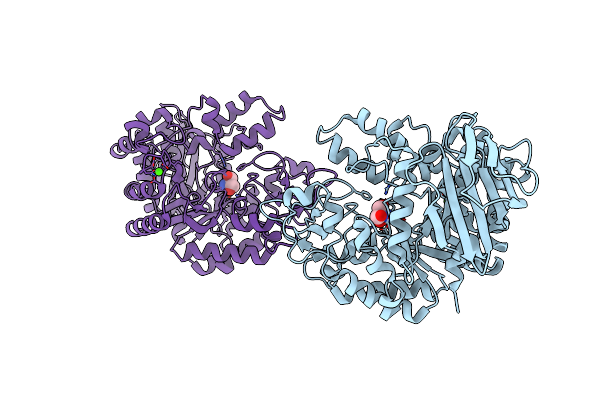
Crystal Structure Of The Trehalulose Synthase Mutb, Mutant A258V, In Complex With Tris
Organism: Pseudomonas mesoacidophila
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-08-21 Classification: ISOMERASE Ligands: CA, TRS |
 |
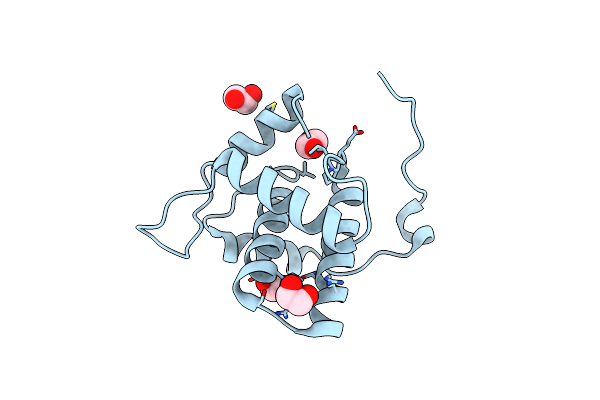
Crystal Structure Of The Trehalulose Synthase Mutant, Mutb D415N, In Complex With Tris
Organism: Pseudomonas mesoacidophila
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-08-21 Classification: ISOMERASE Ligands: CA, TRS |
 |
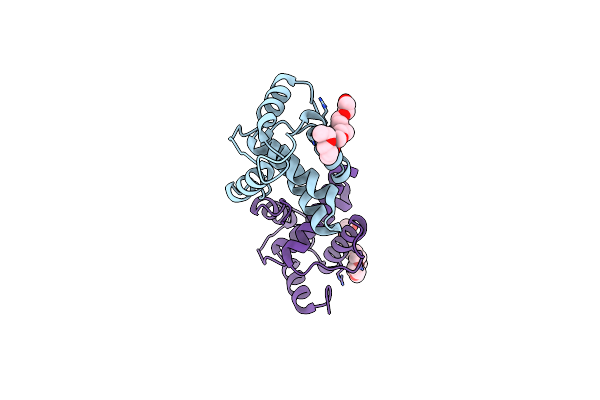
Crystal Structure Of The Full-Length Matrix Subunit (P15) Of The Feline Immunodeficiency Virus (Fiv) Gag Polyprotein
Organism: Feline immunodeficiency virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-10 Classification: VIRAL PROTEIN Ligands: EDO |
 |
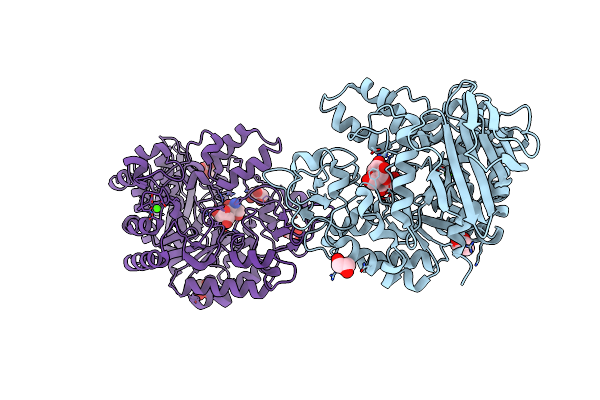
Crystal Structure Of A C-Terminal Truncated Form Of The Matrix Subunit (P15) Of Feline Immunodeficiency Virus (Fiv)
Organism: Feline immunodeficiency virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-10 Classification: VIRAL PROTEIN Ligands: 7PE |
 |
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: CA, GLC, GOL, TRS |
 |
Crystal Structure Of The Mutb F164L Mutant From Crystals Soaked With The Substrate Sucrose
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: CA, GOL, TRS |
 |
Crystal Structure Of The Mutb F164L Mutant From Crystals Soaked With Trehalulose
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: CA, TRS, GOL |
 |
Crystal Structure Of The Mutb F164L Mutant From Crystals Soaked With Isomaltulose
Organism: Rhizobium
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: CA, TRS, GOL |

