Search Count: 13
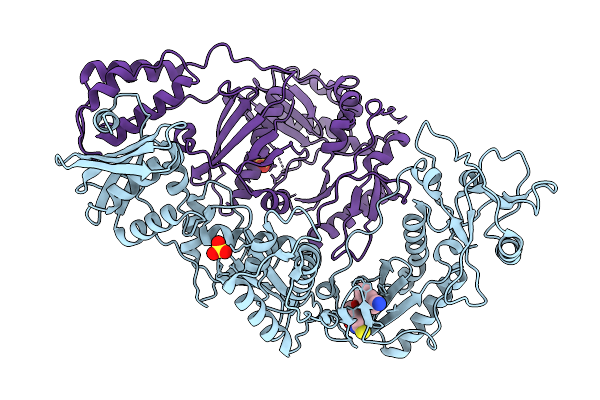 |
Crystal Structure Of Wt Hiv-1 Reverse Transcriptase In Complex With 5-{2-[2-(2-Oxo-4-Sulfanylidene-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy] Phenoxy}Naphthalene-2-Carbonitrile (Jlj648), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: CL, SO4, A1BTU |
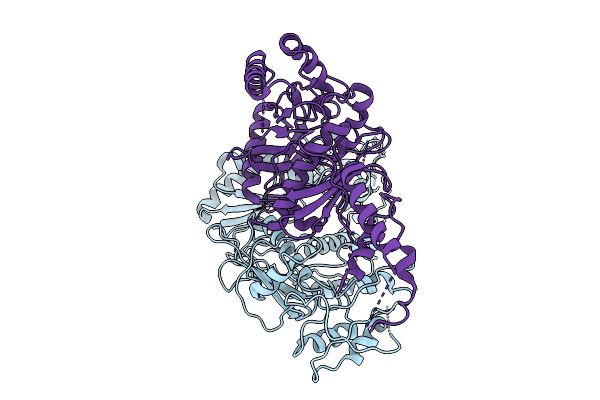 |
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN |
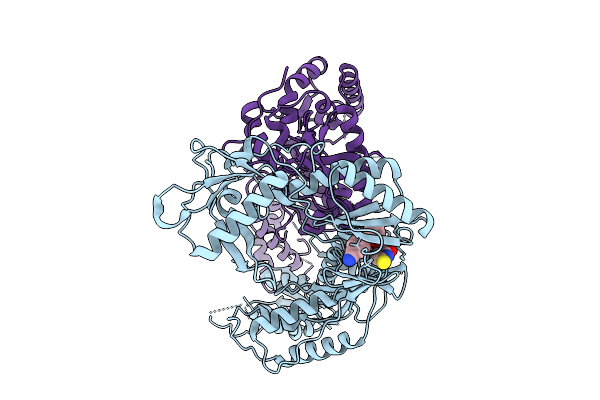 |
Cryo-Em Structure Of Hiv-1 Reverse Transcriptase P66 Homodimer In Complex With 5-{2-[2-(2-Oxo-4-Sulfanylidene-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy] Phenoxy}Naphthalene-2-Carbonitrile (Jlj648), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: A1BTU |
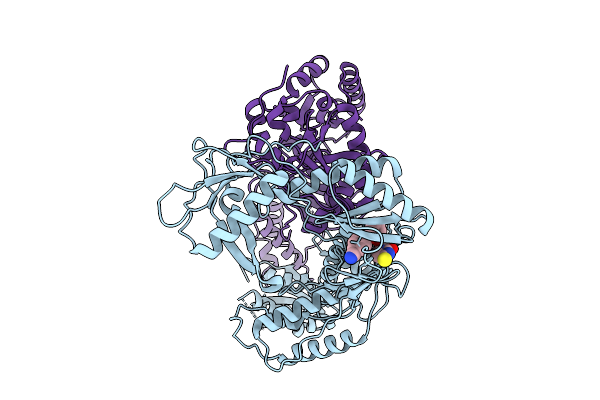 |
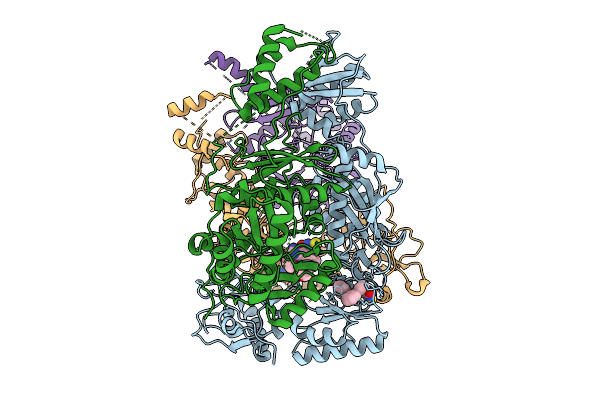
Cryo-Em Structure Of Hiv-1 Reverse Transcriptase P66 Homodimer In Complex With 5-{2-[2-(2-Oxo-4-Sulfanylidene-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy] Phenoxy}Naphthalene-2-Carbonitrile, A Non-Nucleoside Inhibitor, With An Additional Pocket Of Density
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: A1BTU |
 |
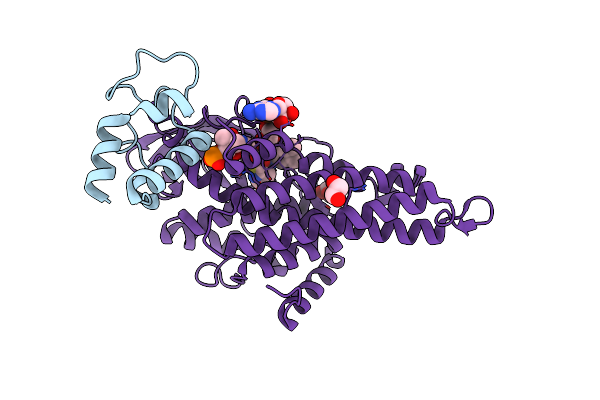
Cryo-Em Structure Of Hiv-1 Reverse Transcriptase P66 Tetramer In Complex With 5-{2-[2-(2-Oxo-4-Sulfanylidene-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy]Phenoxy}Naphthalene-2-Carbonitrile (Jlj648), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: A1BTU |
 |
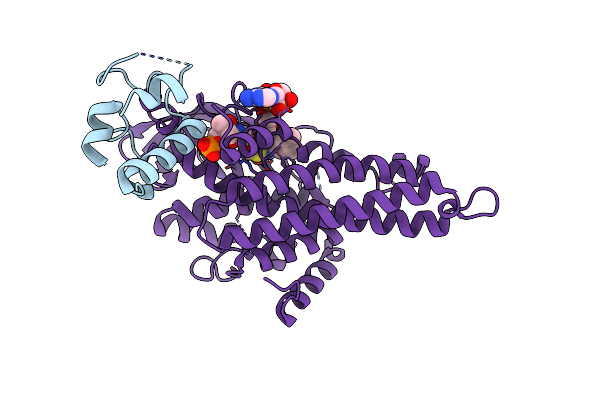
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE Ligands: FK4, FAD, EDO |
 |
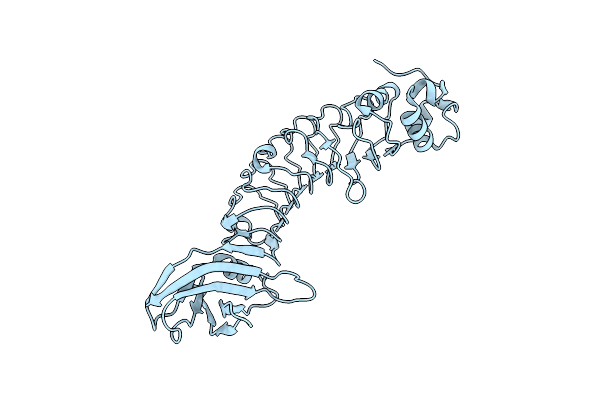
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, PNS |
 |
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-07-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-18 Classification: UNKNOWN FUNCTION Ligands: CA, CL |
 |
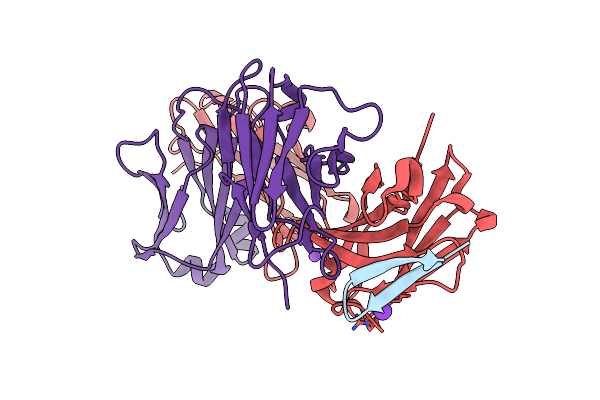
Structure Of The Hepatitis C Virus Envelope Glycoprotein E2 Antigenic Region 412-423 Bound To The Broadly Neutralizing Antibody Ap33
Organism: Homo sapiens, Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-09-26 Classification: IMMUNE SYSTEM |
 |
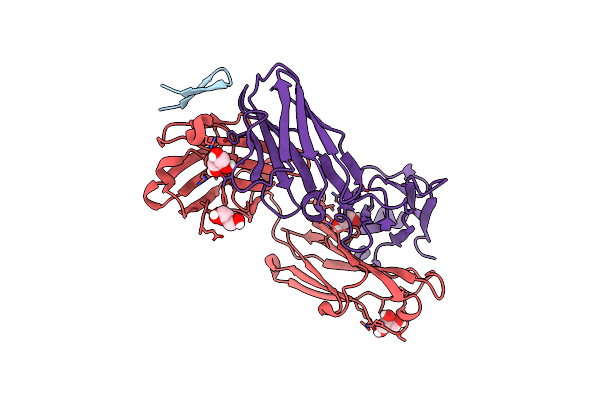
Structure Of The Hepatitis C Virus Envelope Glycoprotein E2 Antigenic Region 412-423 Bound To The Broadly Neutralizing Antibody Hcv1, P2(1) Form
Organism: Homo sapiens, Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2012-05-23 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Structure Of The Hepatitis C Virus Envelope Glycoprotein E2 Antigenic Region 412-423 Bound To The Broadly Neutralizing Antibody Hcv1, C2 Form
Organism: Homo sapiens, Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-05-23 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: GOL, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1995-01-26 Classification: COMPLEX(TRANSFERASE/PEPTIDE) |

