Search Count: 44
 |
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-01 Classification: HYDROLASE |
 |
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Oxacillin) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 1S6 |
 |
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Cephalexin) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 63U |
 |
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Ertapenem) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 1RG |
 |
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE |
 |
Structure Of The Exo-L-Galactose-6-Sulfatase Bus1_11 From Bacteroides Uniformis
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, NI |
 |
Structure Of Exo-L-Galactose-6-Sulfatase Bus1_11 From Bacteroides Uniformis In Complex With Neoporphyrabiose
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, NA |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: EDO, CA |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius In Complex With L-Galactose
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, GXL, EDO |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 (D264N Mutant) From Bacteroides Plebeius In Complex With Paranitrophenyl-Alpha-L-Galactopyranoside
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, Y9Y, NI, NA, CL |
 |
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-09-16 Classification: PROTEIN TRANSPORT Ligands: CD, CL, NA |
 |
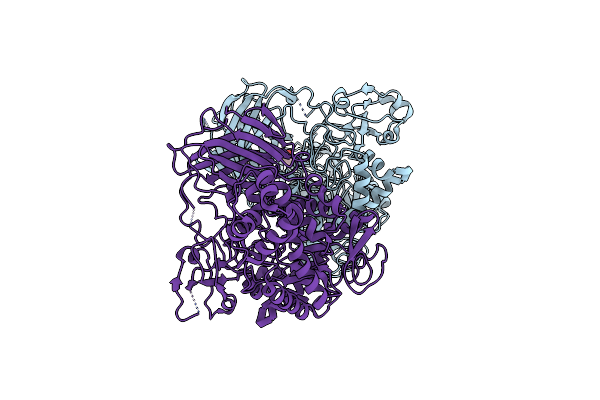
Crystal Structure Of The Type Iii Secretion System Pilotin-Secretin Complex Invh-Invg
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-16 Classification: PROTEIN TRANSPORT Ligands: SO4 |
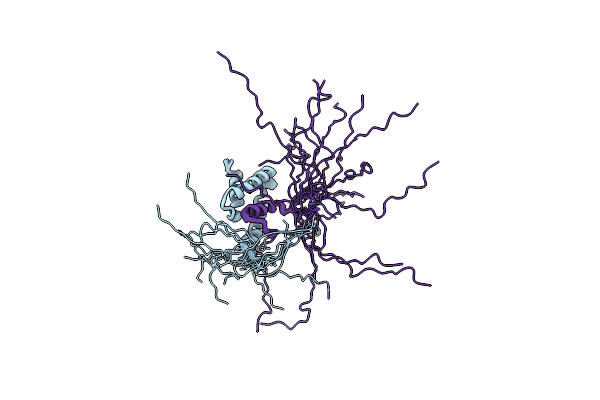 |
Structural Characterization Of The Type Iii Secretion System Pilotin-Secretin Complex Invh-Invg By Nmr Spectroscopy
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: SOLUTION NMR Release Date: 2020-09-16 Classification: PROTEIN TRANSPORT |
 |
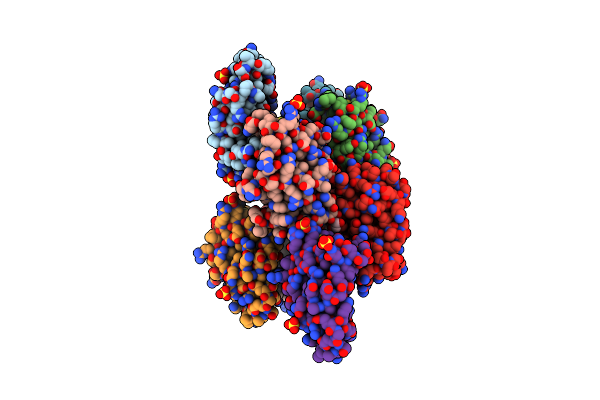
Organism: Pseudoalteromonas fuliginea
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: EDO |
 |
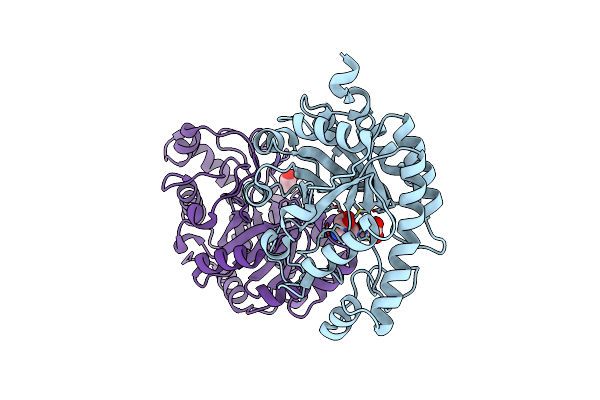
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-06 Classification: PROTEIN BINDING Ligands: SO4, PGE |
 |
Organism: Muricauda sp. mar_2010_75
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: TRS, GOL |
 |
Organism: Muricauda sp. mar_2010_75
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: TRS |
 |
Organism: Formosa agariphila kmm 3901
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: BR, MG |
 |
Crystal Structure Of The Family S1_7 Ulvan-Specific Sulfatase Fa22070 From Formosa Agariphila
Organism: Formosa agariphila
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: CA |
 |
Organism: Formosa agariphila
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2019-06-26 Classification: ISOMERASE |

