Search Count: 42
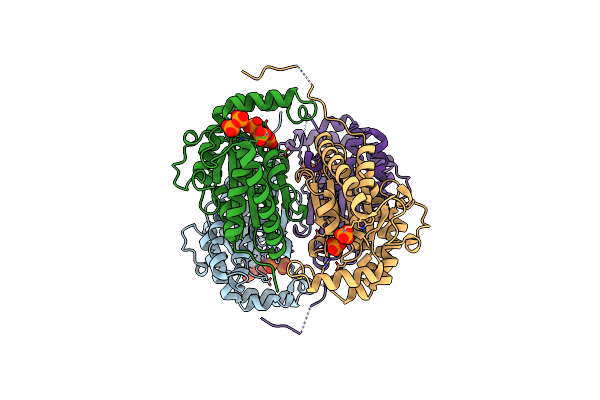 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: PO4, DPO |
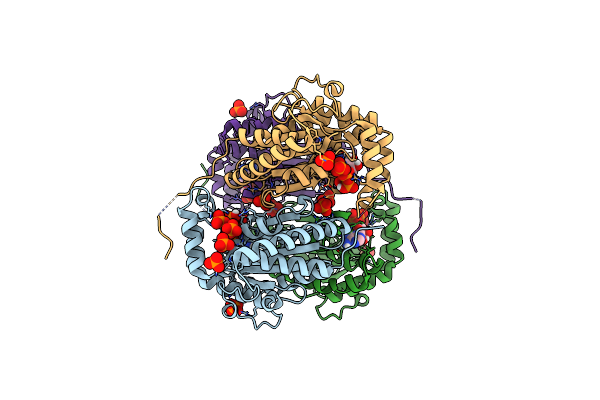 |
Structure Of Polyphosphate Kinase From Meiothermus Ruber N121D Bound To Atp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: ATP, PO4 |
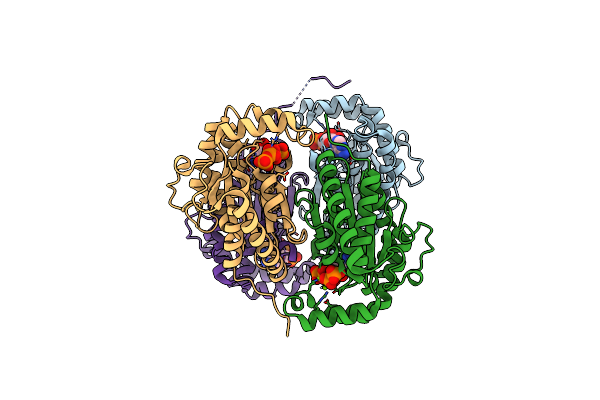 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp And Ppi
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ADP, PPV, MG |
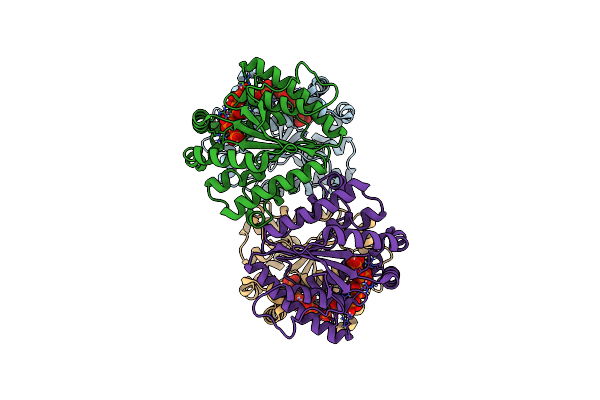 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis Schu S4 With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: 9PI |
 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis With Amppch2Ppp And Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: CL, MG, 6YZ, 6YW |
 |
Structure Of Polyphosphate Kinase 2 Mutant D117N From Francisella Tularensis With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: PO4, 6YX, 6YY, CL, 6YW |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: PO4, SO4 |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: AMP, PO4, SO4, MG |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Atp
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ATP, MG, SO4, GOL |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ADP, PO4, MG, GOL, CL |
 |
Characterisation Of Polyphosphate Kinase 2 From The Intracellular Pathogen Francisella Tularensis
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2015-12-02 Classification: TRANSFERASE |
 |
Organism: Thermoanaerobacter italicus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2015-02-04 Classification: LYASE Ligands: SF4, MET, FE, ALA, H2S, NA, SAM |
 |
Crystal Structure Of Thermosynechococcus Elongatus Lipoyl Synthase 2 Complexed With Mta And Dtt
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-08-20 Classification: TRANSFERASE Ligands: SF4, DTT, MTA |
 |
The Crystal Structure Of Lipoyl Synthase In Complex With S-Adenosyl Homocysteine
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Release Date: 2014-08-20 Classification: TRANSFERASE Ligands: SF4, SAH, NA |
 |
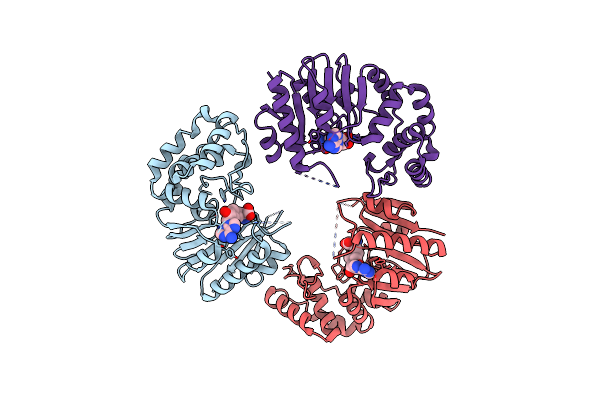
Crystal Structure Of E. Coli Dna Adenine Methyltransferase In Complex With Sah
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2014-02-26 Classification: TRANSFERASE Ligands: SAH |
 |
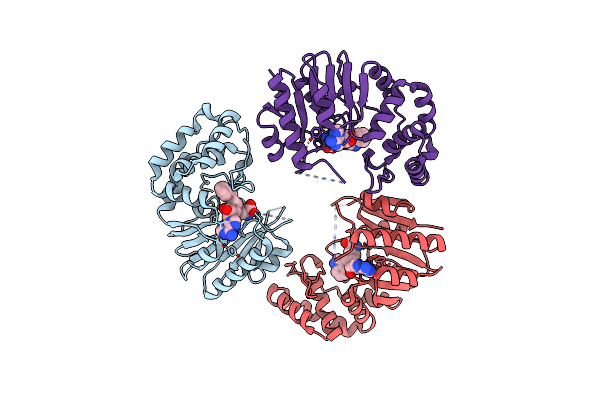
Crystal Structure Of E. Coli Dna Adenine Methyltransferase In Complex With Methylated Aza-Sam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2014-02-26 Classification: TRANSFERASE Ligands: SA8 |
 |
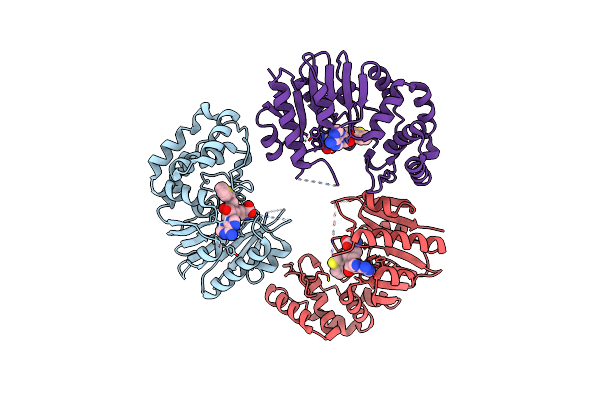
Crystal Structure Of E. Coli Dna Adenine Methyltransferase In Complex With Aza-Sam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-02-26 Classification: TRANSFERASE Ligands: 0Y0 |
 |
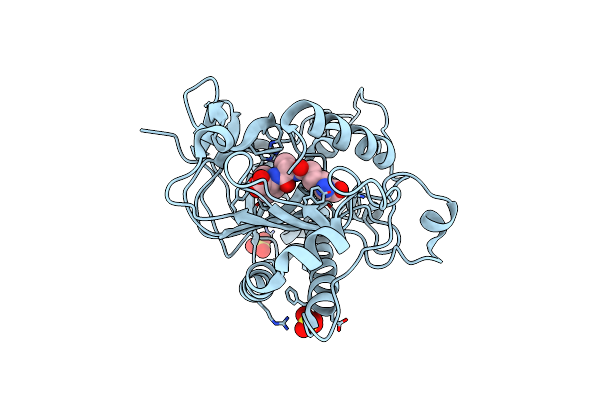
Crystal Structure Of E. Coli Dna Adenine Methyltransferase In Complex With Indole Aza-Sam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2014-02-26 Classification: TRANSFERASE Ligands: 0Y1 |
 |
Crystal Structure Of E. Coli Dna Adenine Methyltransferase In Complex With Benzothiophene Aza-Sam
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-02-26 Classification: TRANSFERASE Ligands: 0Y2 |
 |
Isopenicillin N Synthase From Aspergillus Nidulans (Anaerobic Ac- Methyl-Cyclopropylglycine Fe Complex)
Organism: Emericella nidulans (strain fgsc a4 / atcc 38163 / cbs 112.46 / nrrl 194 / m139)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2007-04-10 Classification: OXIDOREDUCTASE Ligands: ACW, SO4, FE2 |

