Search Count: 18
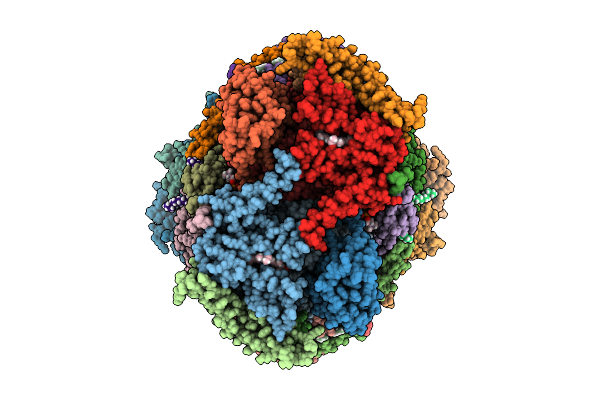 |
Toxoplasma Gondii Cytochrome Bc1 Complex From The Respiratory Supercomplex Iii2-Iv Inhibited By Atovaquone And Elq-300
Organism: Toxoplasma gondii gt1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: ELECTRON TRANSPORT Ligands: CDL, AOQ, A1IJD, HEM, HEC, FES, PC1, ZN |
 |
Cryo-Em Structure Of The Atovaquone-Inhibited Complex Iii From The Chlorocebus Sabaeus Respirasome
Organism: Chlorocebus sabaeus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ELECTRON TRANSPORT Ligands: HEC, ZN, FES, CDL, PC1, AOQ, PEE, AFI, HEM |
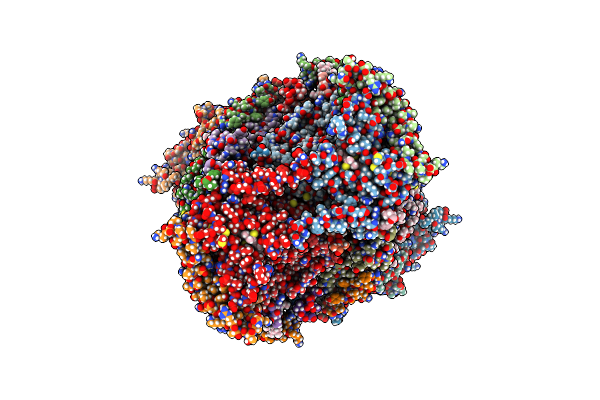 |
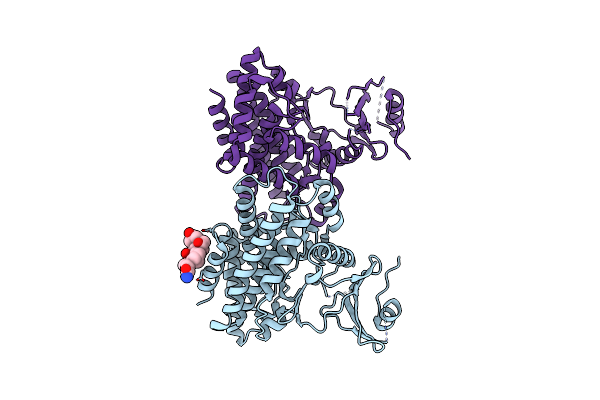
Cryo-Em Structure Of The Toxoplasma Gondii Respiratory Chain Complex Iii Inhibited By Elq-300
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ELECTRON TRANSPORT Ligands: CDL, A1IJD, HEM, MG, HEC, PC1, ZN |
 |
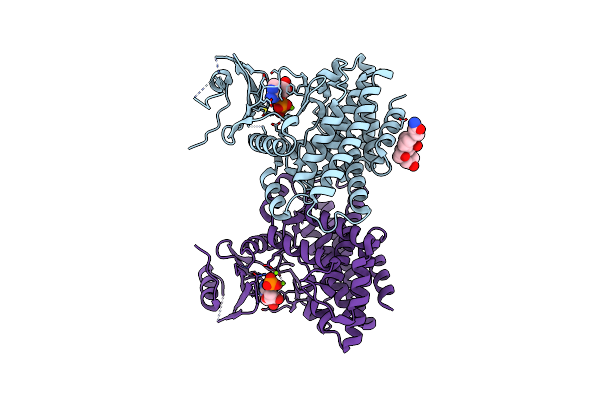
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: CPS |
 |
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, ADP, CPS |
 |
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, CPS, ACP |
 |
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, CPS, ACP, SR1 |
 |
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, ADP, PO4, CPS |
 |
Crystal Structure Of Mta/Adohcy Nucleosidase Glu12Gln Mutant Complexed With 5-Methylthioribose And Adenine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-10-04 Classification: HYDROLASE Ligands: SR1, ADE |
 |
Crystal Structure Of Mta/Adohcy Nucleosidase Asp197Asn Mutant Complexed With 5'-Methylthioadenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-10-04 Classification: HYDROLASE Ligands: MTA |
 |
Crystal Structure Of Mta/Adohcy Nucleosidase With A Ligand-Free Purine Binding Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-10-04 Classification: HYDROLASE Ligands: PE5, GOL, IPA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-03-01 Classification: HYDROLASE Ligands: CL, TDI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-03-01 Classification: HYDROLASE Ligands: MTM |
 |
Structure Of Human 5'-Deoxy-5'-Methylthioadenosine Phosphorylase Complexed With Formycin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2004-05-18 Classification: TRANSFERASE Ligands: FMC |
 |
Structure Of Human 5'-Deoxy-5'-Methylthioadenosine Phosphorylase Complexed With 5'-Methylthiotubercidin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-05-18 Classification: TRANSFERASE Ligands: SO4, MTH |
 |
Crystal Structure Of E. Coli Mta/Adohcy Nucleosidase Complexed With 5'-Methylthiotubercidin (Mth)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-11-25 Classification: HYDROLASE Ligands: MTH |
 |
Crystal Structure Of E. Coli Mta/Adohcy Nucleosidase Complexed With Formycin A (Fma)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-03-25 Classification: HYDROLASE Ligands: FMC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-10-01 Classification: HYDROLASE Ligands: ADE |

