Search Count: 268
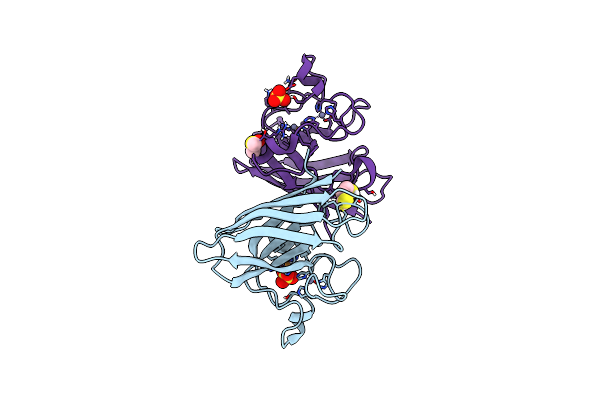 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: CU, ZN, SO4, UDI, DMS |
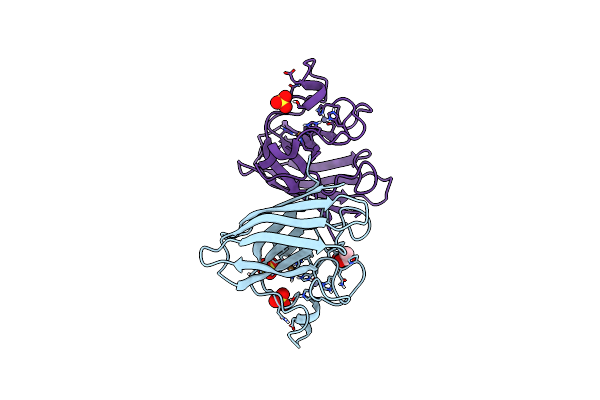 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: ACT, ZN, CU, SO4 |
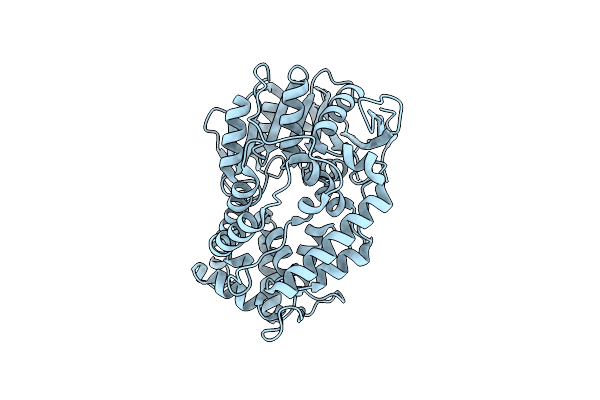 |
Crystal Structure Of Hip1 (Rv2224C) Mutant - T466A/S228Dha (Dehydroalanine)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-30 Classification: HYDROLASE |
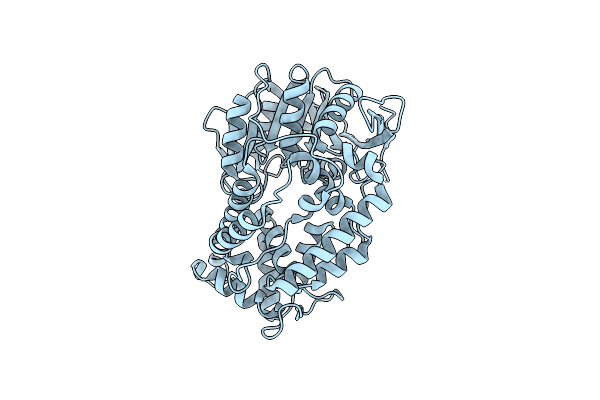 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-03-23 Classification: HYDROLASE |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: SO4, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Catalytic Core
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-04-25 Classification: LYASE Ligands: PLP, ACT, CA, NA, CL, PEG, PGE, EDO |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Plp-L-Serine Intermediate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-04-25 Classification: LYASE Ligands: EVM, CA, NA, CL, PEG, PGE, EDO |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Plp-Aminoacrylate Intermediate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2018-04-25 Classification: LYASE Ligands: P1T, CA, NA, CL, PEG, PGE, EDO |

