Search Count: 213
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: SIGNALING PROTEIN |
 |
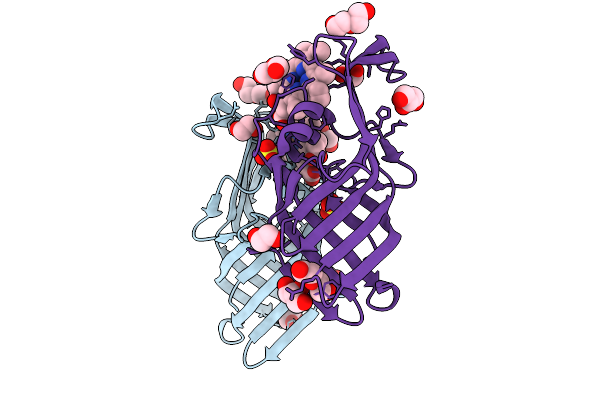
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: HEM, PEG, EDO, GOL |
 |
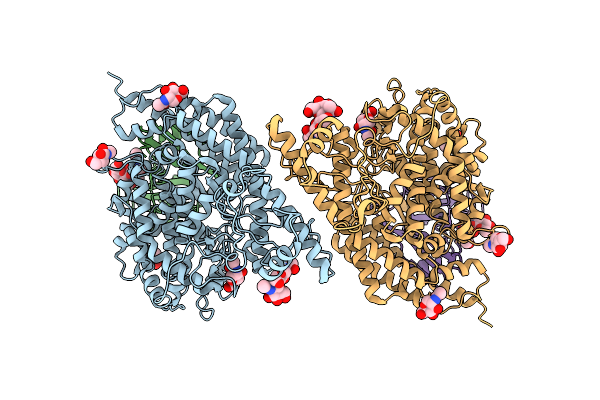
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: HEM, GOL, PEG, EDO, SO4 |
 |
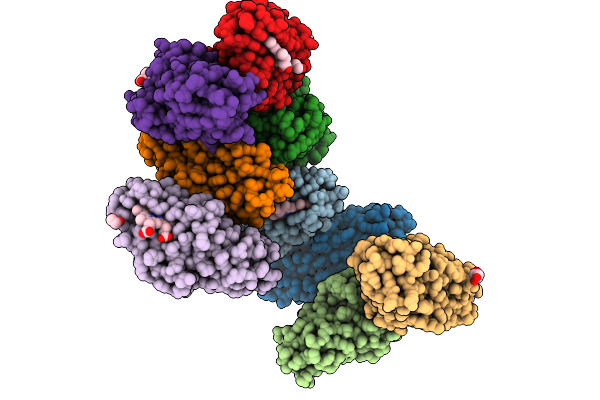
Structure Of Hku5 Spike C-Terminal Domain In Complex With Ace2 From Pipistrellus Abramus
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of Pentameric Outer Membrane Protein A From Bdellovibrio Bacteriovorus
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LIPID TRANSPORT Ligands: PLM, OCT, BGC, OC9 |
 |
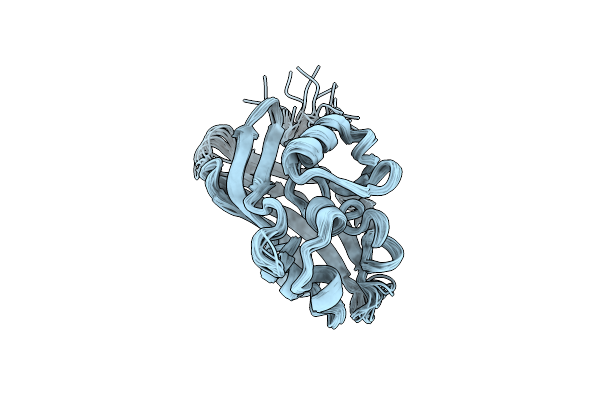
Cryo-Em Structure Of Pentameric Outer Membrane Protein A From Bdellovibrio Bacteriovorus
Organism: Bdellovibrio bacteriovorus hd100
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: LIPID BINDING PROTEIN |
 |
Solution Structure Of The Hemoglobin Receptor Hbpa From Corynebacterium Diphtheriae
Organism: Corynebacterium diphtheriae nctc 13129
Method: SOLUTION NMR Release Date: 2025-01-08 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Human Hemoglobin In Complex With The Hbpa Receptor From Corynebacterium Diphtheriae
Organism: Corynebacterium diphtheriae nctc 13129, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-01-08 Classification: PROTEIN BINDING Ligands: HEM, GOL, PO4 |
 |
Organism: Neobacillus niacini
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: FAD, DR9 |
 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, DR9 |
 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, WTQ, DR9 |
 |
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2024-04-24 Classification: SIGNALING PROTEIN |
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: MEMBRANE PROTEIN Ligands: TWT, PEE, R16, CLR, NAG, CA, PLM |
 |
Structure Of The C-Terminal Beta Helix Domain Of The Bdellovibrio Bacteriovorus Bd3182 Fibre
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-10-25 Classification: CELL ADHESION |
 |
Structure Of The C-Terminal Domain Of The Bdellovibrio Bacteriovorus Bd2133 Fibre
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-25 Classification: CELL ADHESION Ligands: SO4, NA, CL |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-10-25 Classification: CELL ADHESION |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-10-25 Classification: CELL ADHESION |
 |
Structure Of The C-Terminal Domains Of The Bdellovibrio Bacteriovorus Bd2439 Fibre In Complex With Glcnac
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-10-25 Classification: CELL ADHESION Ligands: IOD, PG4, PEG, NAG, SO4, B3P |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2023-10-25 Classification: CELL ADHESION Ligands: PO4 |

