Search Count: 16
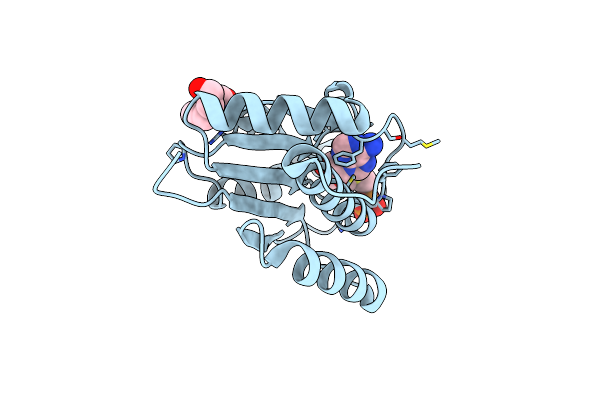 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-06-06 Classification: TRANSFERASE, RIBOSOMAL PROTEIN Ligands: SAM, PG4, PO4 |
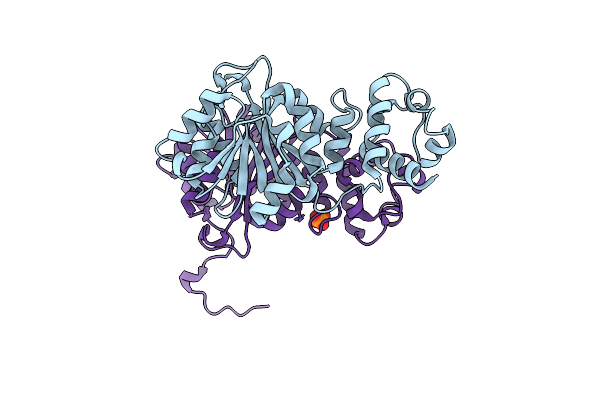 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-15 Classification: TRANSFERASE Ligands: CL, PO4 |
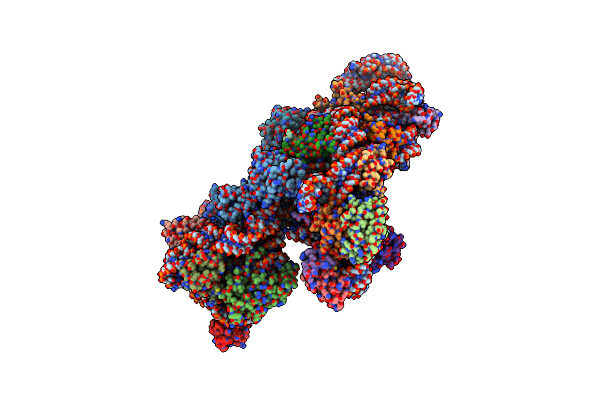 |
Structure Of The E. Coli Methyltransferase Ksga Bound To The E. Coli 30S Ribosomal Subunit
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:13.50 Å Release Date: 2012-02-15 Classification: TRANSLATION |
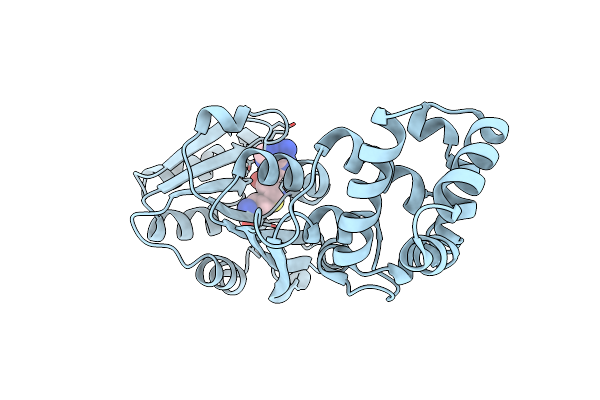 |
Crystal Structure Of The Complex Between S-Adenosyl Homocysteine And Methanocaldococcus Jannaschi Dim1.
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The Complex Between Amp And Methanocaldococcus Jannaschi Dim1
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: AMP, SO4 |
 |
Crystal Structure Of The Complex Between Adenosine And Methanocaldococcus Jannaschi Dim1
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: ADN, SO4 |
 |
Crystal Structure Of The Complex Between S-Adenosyl Methionine And Methanocaldococcus Jannaschi Dim1.
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: SAM, PO4 |
 |
Crystal Structure Of Dim1 From The Thermophilic Archeon, Methanocaldococcus Jannaschi
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-06-30 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Dim1 From The Thermophilic Archeon, Methanocaldococcus Jannaschi
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-06-30 Classification: TRANSFERASE Ligands: SO4 |
 |
2.1 Angstrom Crystal Structure Of Ksga: A Universally Conserved Adenosine Dimethyltransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-06-29 Classification: TRANSFERASE, TRANSLATION |
 |
Method: X-RAY DIFFRACTION
Resolution:1.80 Å Release Date: 2003-03-04 Classification: RNA Ligands: TL |
 |
Method: X-RAY DIFFRACTION
Resolution:1.85 Å Release Date: 2003-03-04 Classification: RNA Ligands: BA |
 |
 |
 |
Helix 45 (16S Rrna) From B. Stearothermophilus, Nmr, Minimized Average Structure
|
 |

