Search Count: 19
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW9 |
 |
Crystal Structure Of The Mertk Kinase Domain In Complex With Inhibitor Mips15692
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2021-10-06 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: YR7 |
 |
K. Pneumoniae Topoisomerase Iv (Pare-Parc) In Complex With Dna And Compound 34 (7-[(1S,5R)-1-Amino-3-Azabicyclo[3.1.0]Hexan-3-Yl]-4-(Aminomethyl)-1-Cyclopropyl-3,6-Difluoro-8-Methylquinolin-2(1H)-One)
Organism: Klebsiella pneumoniae 342
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-07-15 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: MPD, MG, CL, TNJ |
 |
Braf In Complex With N-(4-Methyl-3-(1-Methyl-2-Oxo-2,3-Dihydro-1H-Benzo[D]Imidazol-5-Yl)Phenyl)-3-(Trifluoromethyl)Benzamide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-10-23 Classification: transferase/transferase inhibitor Ligands: K7S |
 |
Braf In Complex With N-(3-(2-(2-Hydroxyethoxy)-6-Morpholinopyridin-4-Yl)-4-Methylphenyl)-2-(Trifluoromethyl)Isonicotinamide (Lxh254)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2019-09-04 Classification: transferase/transferase inhibitor Ligands: K81 |
 |
Solution Nmr Structure Of The Major Type Iv Pilin Ppdd From Enterohemorrhagic Escherichia Coli (Ehec)
Organism: Escherichia coli o157:h7
Method: SOLUTION NMR Release Date: 2019-05-15 Classification: CELL ADHESION |
 |
Structure Of The Type Iv Pilus From Enterohemorrhagic Escherichia Coli (Ehec)
Organism: Escherichia coli o157:h7
Method: ELECTRON MICROSCOPY Release Date: 2019-05-15 Classification: PROTEIN FIBRIL |
 |
Chain A. Udp-3-O-[3-Hydroxymyristoyl] N-Acetylglucosamine Deacetylase Pa-Lpxc Complexed With (R)-3-((S)-3-(4-(Cyclopropylethynyl)Phenyl)-2-Oxooxazolidin-5-Yl)-N-Hydroxy-2-Methyl-2-(Methylsulfonyl)Propenamide
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: JBA, ZN |
 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-01-31 Classification: PROTEIN FIBRIL |
 |
Braf In Complex With N-(3-(Tert-Butyl)Phenyl)-4-Methyl-3-(6-Morpholinopyrimidin-4-Yl)Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2017-06-28 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: 92D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-28 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: 92J |
 |
Crystal Structure Of Hsp90-Alpha N-Domain Bound To The Inhibitor Nvp-Hsp990
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-26 Classification: CHAPERONE/INHIBITOR Ligands: 3JC |
 |
Crystal Structure Of Hsp90-Alpha N-Domain Bound To The Inhibitor Nvp-Hsp990
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-11-19 Classification: CHAPERONE Ligands: 990 |
 |
Structure Of A Designed Meningococcal Antigen (Factor H Binding Protein, Mutant G1) Inducing Broad Protective Immunity
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-27 Classification: IMMUNE SYSTEM |
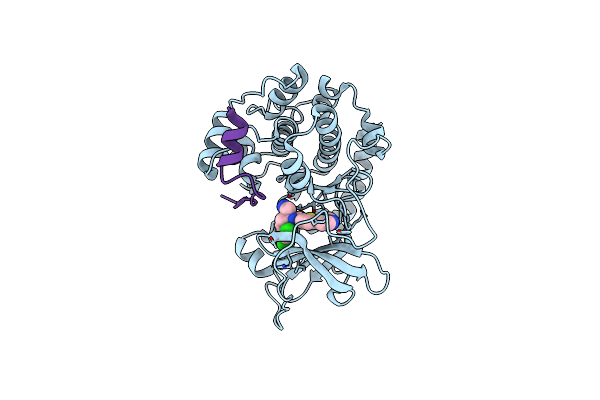 |
Discovery Of 2-Pyrimidyl-5-Amidothiophenes As Novel And Potent Inhibitors For Akt: Synthesis And Sar Studies
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-05-01 Classification: SIGNALING PROTEIN,TRANSFERASE/INHIBITOR Ligands: 796 |
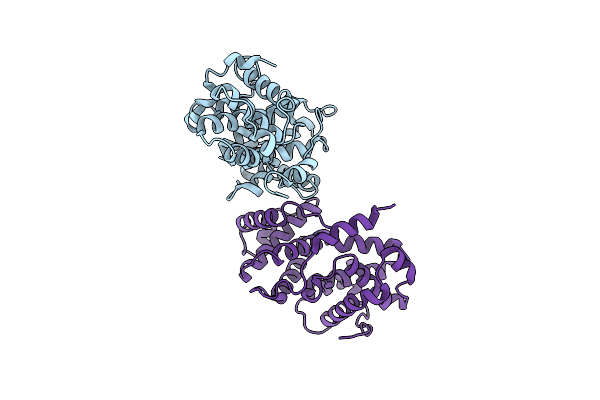 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-10-17 Classification: CELL CYCLE |

