Search Count: 31
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-12-24 Classification: CELL CYCLE Ligands: BEZ, NA |
 |
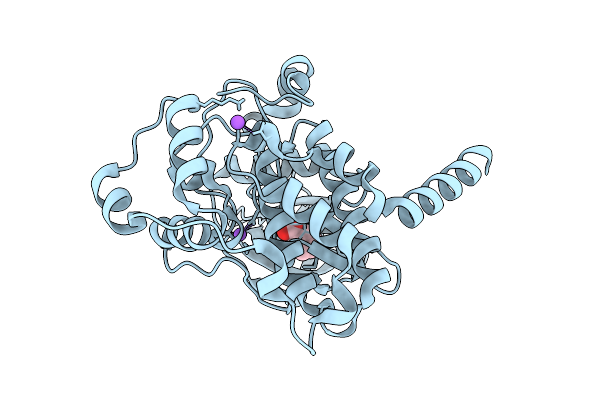
Arabidopsis Thaliana Casein Kinase 2 (Ck2) Alpha1 - Beta1 Complex Bound To Inositol Hexakisphosphate (Insp6)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2025-12-17 Classification: CELL CYCLE Ligands: IHP, ZN |
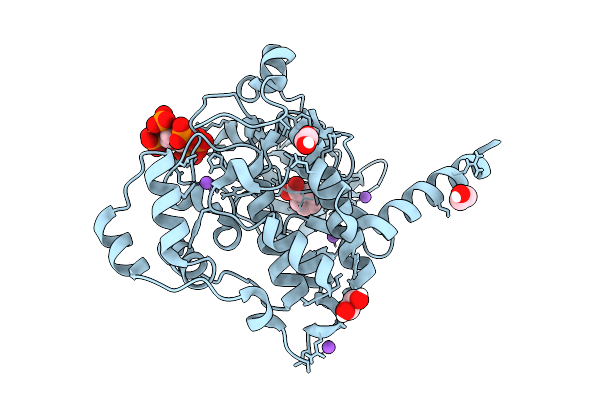 |
Arabidopsis Thaliana Casein Kinase 2 (Ck2) Alpha Isoform 1 In Complex With Inositol Hexakisphosphate (Insp6)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-12-10 Classification: CELL CYCLE Ligands: IHP, BEZ, EDO, CL, NA |
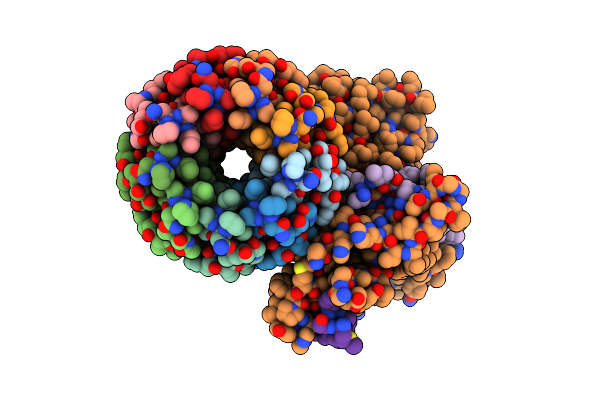 |
Cryo-Em Structure Of Sq31F-Bound Mycobacterium Smegmatis Atp Synthase Fo Region
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: SQC |
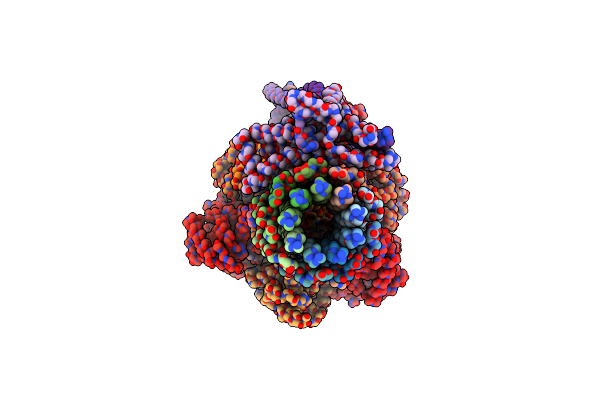 |
Cryo-Em Structure Of Sq31F-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: SQC, ATP, MG, PO4 |
 |
Cryo-Em Structure Of Sq31F-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 2 (Backbone Model)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: ATP, MG, PO4, SQC |
 |
Cryo-Em Structure Of Sq31F-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 3
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: ATP, MG, PO4, SQC |
 |
Cryo-Em Structure Of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Fo Region
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: YGR |
 |
Cryo-Em Structure Of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: ATP, MG, PO4, YGR |
 |
Cryo-Em Structure Of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 2 (Backbone Model)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: ATP, MG, PO4, YGR |
 |
Cryo-Em Structure Of Tbaj-876-Bound Mycobacterium Smegmatis Atp Synthase Rotational State 3
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: TRANSLOCASE/INHIBITOR Ligands: ATP, MG, PO4, YGR |
 |
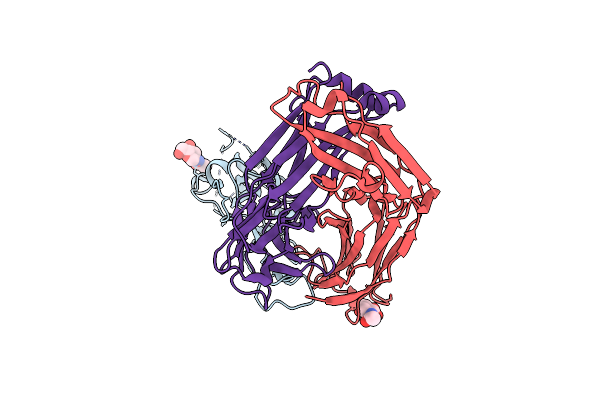
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-270
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-250
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
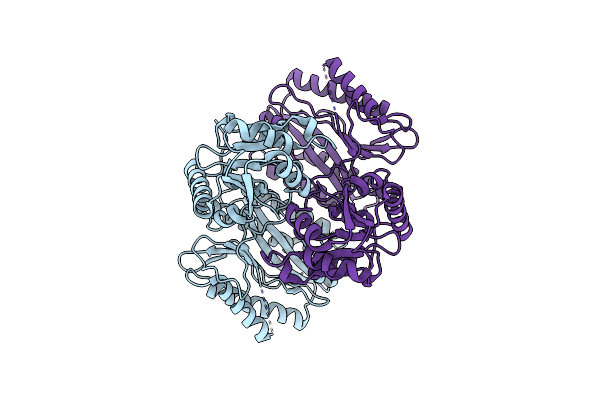 |
Apo Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-20 Classification: HYDROLASE |
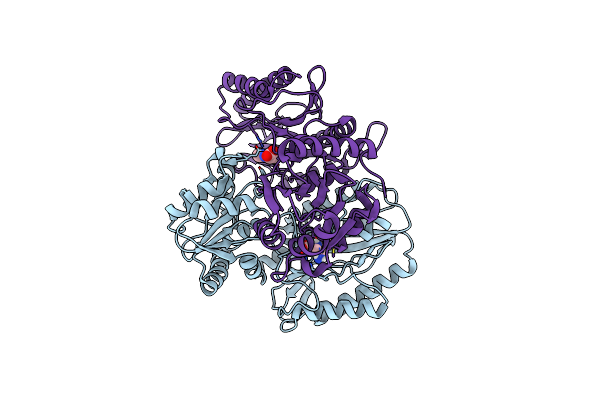 |
Substrate Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: P4B, 4CS |
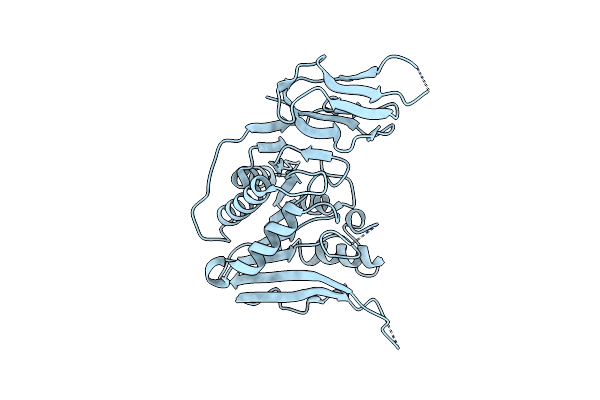 |
Apo Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-20 Classification: HYDROLASE |
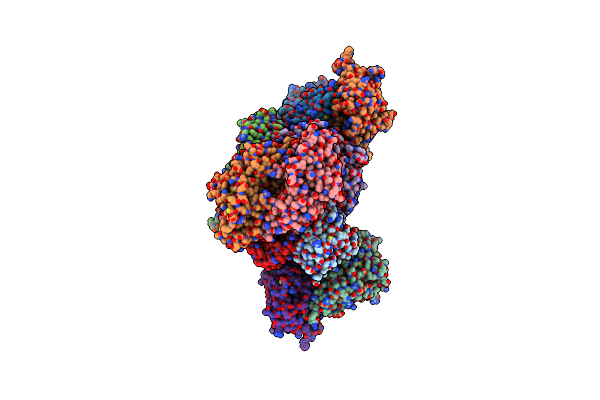 |
Product Bound Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, DAB, ACT |
 |
Product Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, P4B |
 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Its Product Adaba
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: GOL, 9YT, TRS |
 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Coenzyme A
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: COA, ACT |

