Search Count: 67
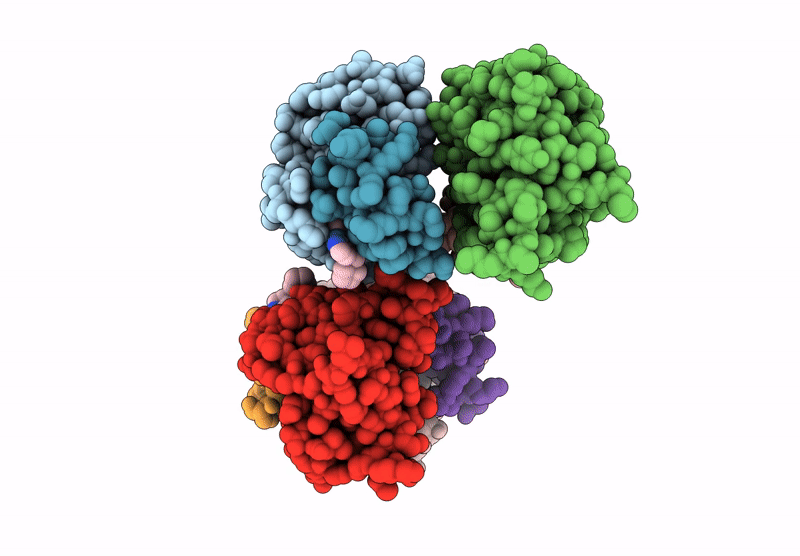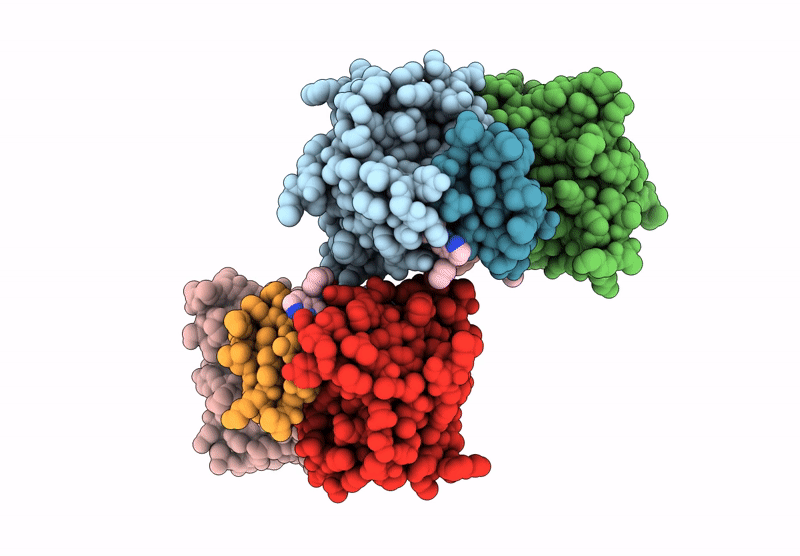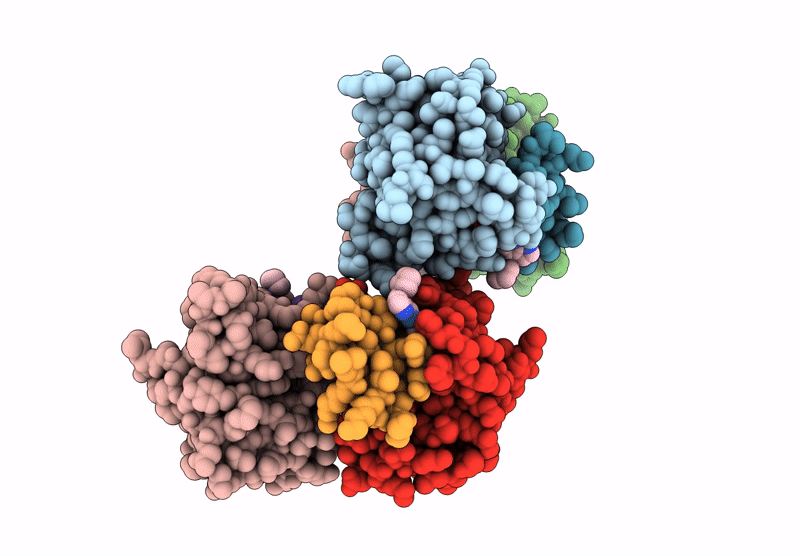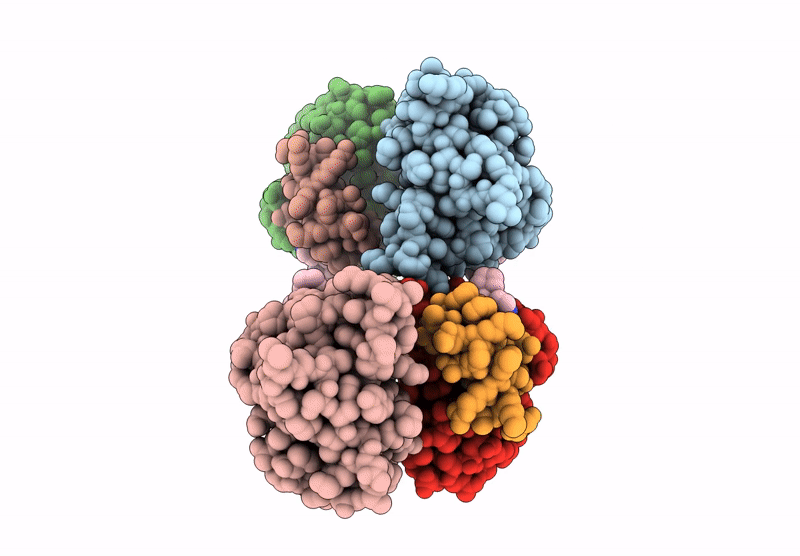 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: IMMUNE SYSTEM Ligands: A1CAC, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: IMMUNE SYSTEM Ligands: A1CAV, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: IMMUNE SYSTEM Ligands: A1CAW, ZN |
 |
Pvtx-405: A Potent, Highly Selective, And Orally Efficacious Molecular Glue Degrader Of Ikzf2 For Cancer Immunotherapy
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: A1A8N, ZN |
 |
Discovery Of Potent, Highly Selective And Efficacious Smarca2 Degraders - Compound 11
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: A1BF5, ZN |
 |
Discovery Of Potent, Highly Selective And Efficacious Smarca2 Degraders - Compound 13
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: A1A1O, ZN, ACT |
 |
Discovery Of Potent, Highly Selective And Efficacious Smarca2 Degraders - Compound 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: A1A1P, ZN, ACT |
 |
Organism: Bat sars-like coronavirus rsshc014
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: VIRAL PROTEIN |
 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-10-23 Classification: VIRAL PROTEIN Ligands: ZN, SAH, EDO, IMD, YDT, CL |
 |
Structure Of The Human Ace2 Receptor In Complex With Antibody Fab Fragment, 05B04
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of A Trapped Pab-Agog/Double-Standed Dna Covalent Intermediate (Dna Containing Adenine Opposite To Lesion)
Organism: Pyrococcus abyssi, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2022-08-10 Classification: HYDROLASE Ligands: MPD |
 |
Crystal Structure Of Pyrococcus Abyssi 8-Oxoguanine Dna Glycosylase (Pabagog) In Complex With Dsdna Containing Cytosine Opposite To 8-Oxog
Organism: Pyrococcus abyssi (strain ge5 / orsay), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-08-03 Classification: HYDROLASE Ligands: MPD |
 |
Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant In Complex With 2-(2-Chloro-5-Phenylthieno[2,3-D]Pyrimidin-4-Yl)Isoindolin-5-Ol And Grip Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-03 Classification: TRANSCRIPTION Ligands: 61Z |
 |
Crystal Structure Of A Trapped Pab-Agog/Double-Standed Dna Covalent Intermediate (Dna Containing Cytosine Opposite To Lesion)
Organism: Pyrococcus abyssi (strain ge5 / orsay), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MPD |
 |
Crystal Structure Of A Trapped Pab-Agog/Double-Standed Dna Covalent Intermediate (Dna Containing Thymine Opposite To Lesion)
Organism: Pyrococcus abyssi (strain ge5 / orsay), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MPD, NA |
 |
Crystal Structure Of Tga-Agog, An 8-Oxoguanine Dna Glycosylase From Thermococcus Gammatolerans
Organism: Thermococcus gammatolerans (strain dsm 15229 / jcm 11827 / ej3)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2022-06-22 Classification: HYDROLASE Ligands: CL, GOL |
 |
Crystal Structure Of A Trapped Pab-Agog/Single-Standed Dna Covalent Intermediate
Organism: Pyrococcus abyssi (strain ge5 / orsay), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-06-22 Classification: HYDROLASE Ligands: PO4, K |
 |
Estrogen Receptor Alpha Ligand Binding Domain Y537S In Complex With 4-(((2-Chloro-5-Phenylthieno[2,3-D]Pyrimidin-4-Yl)Amino)Methyl)Phenol And Grip Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-06-22 Classification: TRANSCRIPTION Ligands: 5VP |
 |
Estrogen Receptor Alpha Ligand Binding Domain Y537S In Complex With 2-Chloro-4-((4-Hydroxybenzyl)Amino)-5-Phenylthieno[2,3-D]Pyrimidin-6-Ol And Grip Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-06-22 Classification: TRANSCRIPTION Ligands: EMY |
 |
Crystal Structure Of Pab-Agog, An 8-Oxoguanine Dna Glycosylase From Pyrococcus Abyssi
Organism: Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, CIT, CL, NA |

