Search Count: 73,678
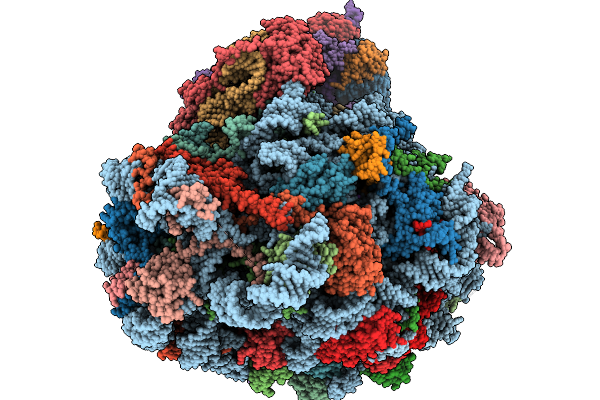 |
Structure Of The Arabidopsis Thaliana 80S Ribosome In Complex With P- And E-Site Trnas And Mrna
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:1.82 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, K, TER, SPD, EPE, ZN |
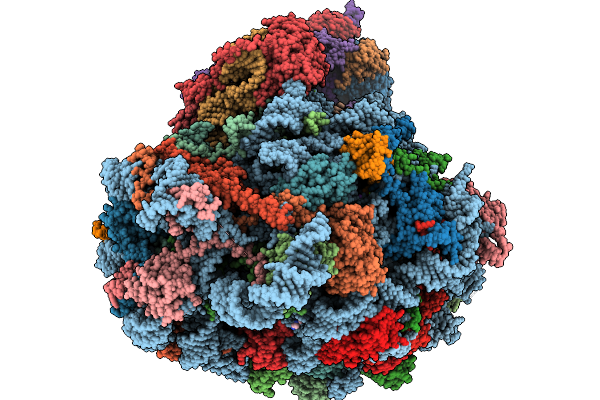 |
Structure Of The Arabidopsis Thaliana 80S Ribosome In Complex With P- And E-Site Trnas, Mrna, And Thermospermine
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: TER, MG, K, SPD, EPE, ZN |
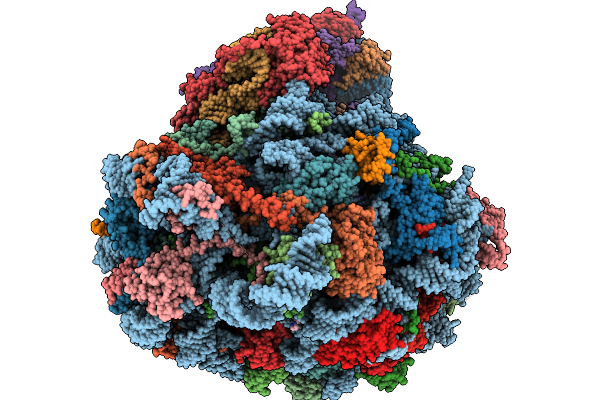 |
Structure Of The Arabidopsis Thaliana 80S Ribosome Ovac Mutant In Complex With P- And E-Site Trnas, Mrna, And Thermospermine
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: TER, MG, K, SPD, EPE, ZN |
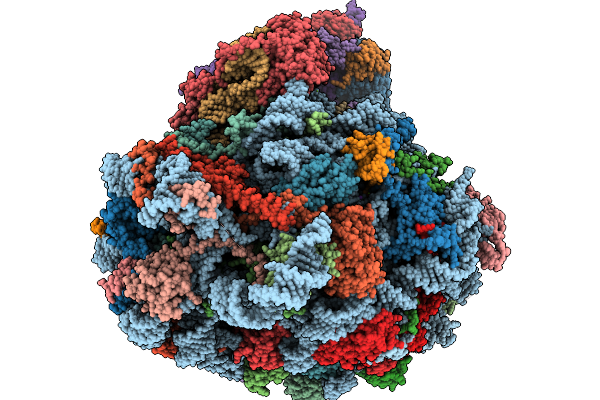 |
Structure Of The Arabidopsis Thaliana 80S Ribosome Ovac Mutant In Complex With P- And E-Site Trnas And Mrna
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, K, TER, SPD, EPE, ZN |
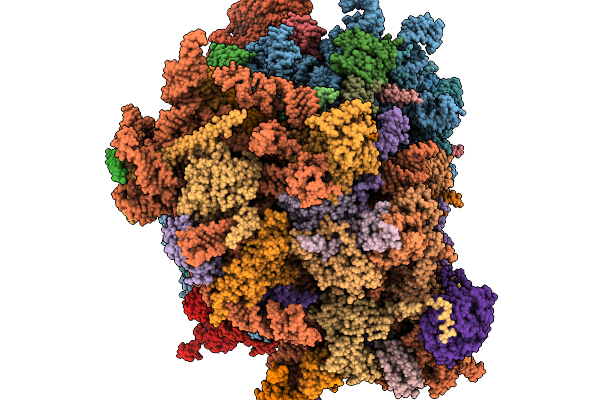 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
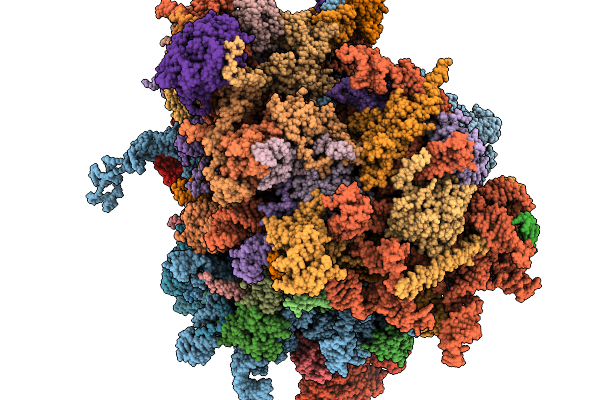 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
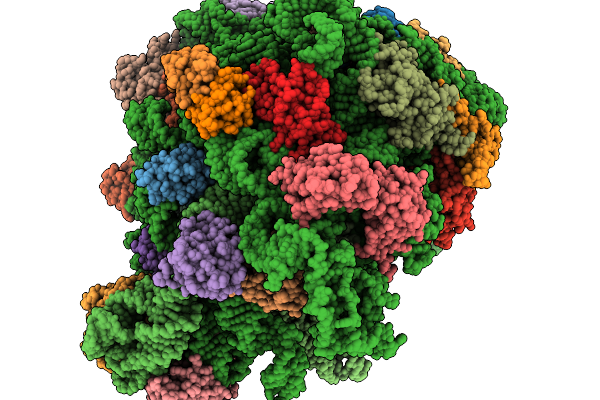 |
Structure Of E.Coli Ribosome With Filamin Mutant Y719E Nascent Chain At Linker Length Of 47 Amino Acids, With Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
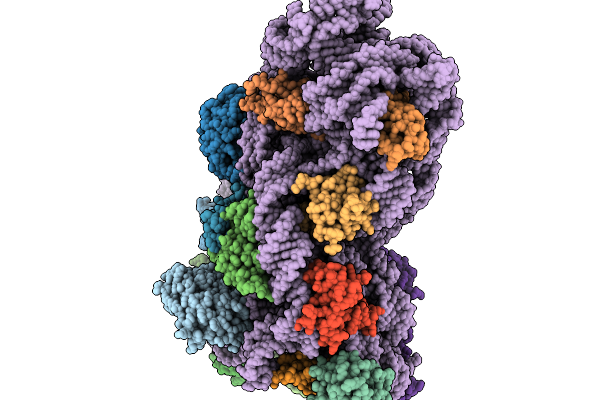 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN |
 |
Structure Of E.Coli Ribosome With Nascent Chain At Linker Length Of 31 Amino Acids, With Mrna, P-Site And A-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: K, MG, NA, SPD, PUT, ZN, HYG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: A1EY1, ECC |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME |
 |
Organism: Escherichia coli, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: ZN, K, MG, PRO |
 |
Cryo-Em Structure Of The Methanosarcina Acetivorans 70S Ribosome In Complex With Sria And Srib
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Resolution:2.03 Å Release Date: 2026-01-28 Classification: RIBOSOME Ligands: MG, ZN |

