Search Count: 22
 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: PHOTOSYNTHESIS Ligands: BCL, BPB, U10, PC1, FE2, SPO, CDL |
 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-05-19 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: OH, HEA, DMU, TRD, HTH, CU, MG, CA, CD, TRS |
 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: TRD, DMU, HEA, CU, MG, CA, HTH, TRS, CD |
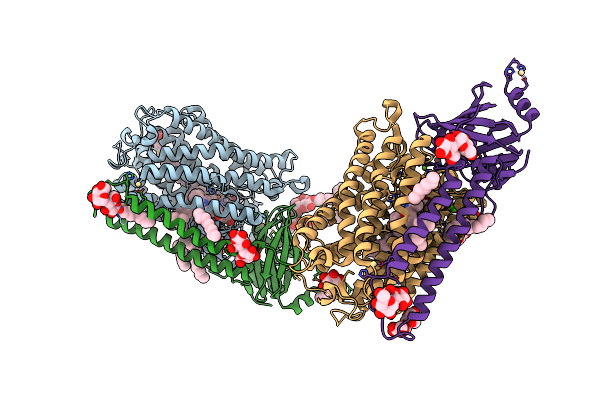 |
Catalytic Core Subunits (I And Ii) Of Cytochrome C Oxidase From Rhodobacter Sphaeroides With K362M Mutation In The Reduced State
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-02-02 Classification: OXIDOREDUCTASE Ligands: DMU, TRD, HEA, CU1, MG, CA, HTH, CD |
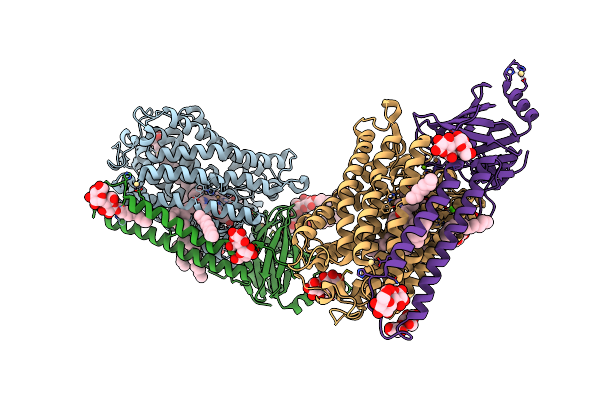 |
Catalytic Core Subunits (I And Ii) Of Cytochrome C Oxidase From Rhodobacter Sphaeroides With K362M Mutation
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-02 Classification: OXIDOREDUCTASE Ligands: OH, DMU, TRD, HEA, CU, MG, CA, HTH, CU1, CD |
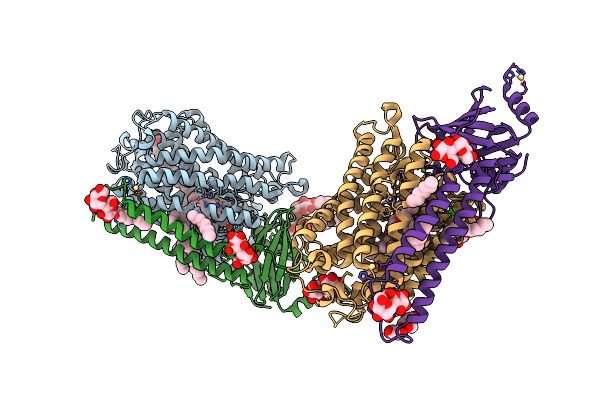 |
Catalytic Core Subunits (I And Ii) Of Cytochrome C Oxidase From Rhodobacter Sphaeroides With D132A Mutation
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2011-02-02 Classification: OXIDOREDUCTASE Ligands: OH, DMU, TRD, HEA, CU, MG, CA, CL, HTH, CU1, CD |
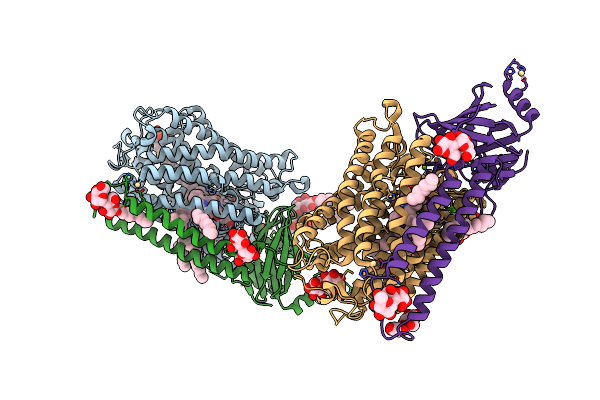 |
Catalytic Core Subunits (I And Ii) Of Cytochrome C Oxidase From Rhodobacter Sphaeroides With D132A Mutation In The Reduced State
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2011-02-02 Classification: OXIDOREDUCTASE Ligands: OH, DMU, TRD, HEA, CU1, MG, CA, CL, HTH, CD |
 |
Crystal Structure Of A Resiniferatoxin-Binding Protein From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-13 Classification: Resiniferatoxin binding protein Ligands: ZN, MG, DTV |
 |
Crystal Structure Of Aminotransferase Prk07036 From Rhodobacter Sphaeroides Kd131
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-07-14 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of The L Protein Of Rhodobacter Sphaeroides Light-Independent Protochlorophyllide Reductase (Bchl) With Mgadp Bound: A Homologue Of The Nitrogenase Fe Protein
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: MG, ADP, SF4 |
 |
Crystal Structure Of Possible 2-Hydroxychromene-2-Carboxylate Isomerase From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-02-03 Classification: ISOMERASE Ligands: GSH, CA, EDO, PGE |
 |
Crystal Structure Of Rhodobacter Sphaeroides Protein Rsp_2168. Northeast Structural Genomics Target Rhr83.
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-12-09 Classification: UNKNOWN FUNCTION |
 |
Solution Structure Of Rhr2 From Rhodobacter Sphaeroides. Northeast Structural Genomics Consortium
Organism: Rhodobacter sphaeroides 2.4.1
Method: SOLUTION NMR Release Date: 2008-08-26 Classification: structural genomics, unknown function |
 |
Crystal Structure Of A Protein Of Unknown Function From Duf427 Family (Rsph17029_0682) From Rhodobacter Sphaeroides 2.4.1 At 2.51 A Resolution
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2008-07-01 Classification: UNKNOWN FUNCTION Ligands: EDO |
 |
Crystal Structure Of Succinylglutamatedesuccinylase/Aspartoacylase From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-03-11 Classification: HYDROLASE Ligands: CA |
 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-01-08 Classification: TRANSCRIPTION |
 |
Solution Nmr Structure Of Rhodobacter Sphaeroides Protein Rhos4_26430. Northeast Structural Genomics Consortium Target Rhr95
Organism: Rhodobacter sphaeroides 2.4.1
Method: SOLUTION NMR Release Date: 2007-10-16 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Crystal Structures Of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter In Its Open Form
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-04-03 Classification: LIGAND BINDING, TRANSPORT PROTEIN Ligands: GOL |
 |
Crystal Structures Of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter In Its Closed Forms
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-04-03 Classification: LIGAND BINDING, TRANSPORT PROTEIN |

