Search Count: 103
 |
F(M197)H Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (100K, 26Kev)
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-07-13 Classification: PHOTOSYNTHESIS Ligands: LDA, UNL, OLC, BCL, BPH, NKP, U10, FE, SPN, EDO |
 |
F(M197)H Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (Room Temperature, 12Kev)
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-07-13 Classification: PHOTOSYNTHESIS Ligands: MYS, LDA, OLC, BCL, BPH, NKP, U10, SPN, FE, EDO |
 |
F(M197)H Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (Room Temperature, 26Kev)
Organism: Rhodobacter sphaeroides (strain atcc 17023 / dsm 158 / jcm 6121 / nbrc 12203 / ncimb 8253 / ath 2.4.1.)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-04-27 Classification: PHOTOSYNTHESIS Ligands: LDA, OLC, MYS, BCL, BPH, NKP, U10, FE, SPN, EDO, PO4 |
 |
Crystal Structure Of R. Sphaeroides Photosynthetic Reaction Center Variant; Y(M210)3-Nitrotyrosine
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-12-29 Classification: PHOTOSYNTHESIS Ligands: LDA, BCL, BPH, U10, CL, FE, SPO, CDL |
 |
Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv In Surfo Crystallization
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: PHOTOSYNTHESIS Ligands: LDA, D12, MYS, PO4, EDO, NA, K, BCL, BPH, U10, D10, DIO, HTO, FE, SPN, CDL |
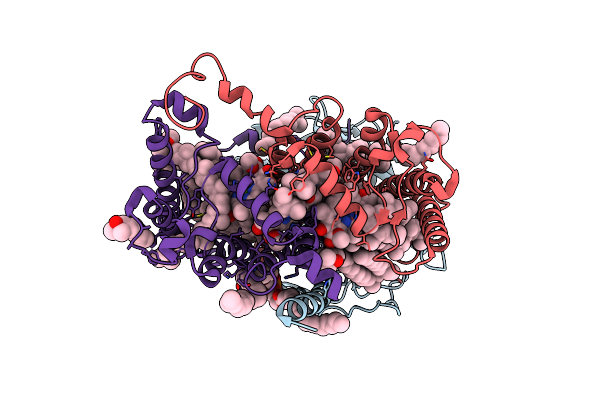 |
Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv Lsp Co-Crystallization With Spheroidene
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: PHOTOSYNTHESIS Ligands: LDA, BCL, BPH, UNL, OLC, FE, U10, SPN, PO4, NKP |
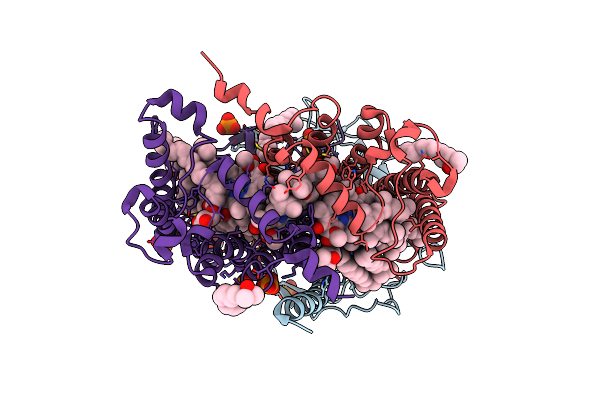 |
Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv Lcp Crystallization
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: PHOTOSYNTHESIS Ligands: LDA, PO4, OLC, BCL, BPH, FE, U10, EDO |
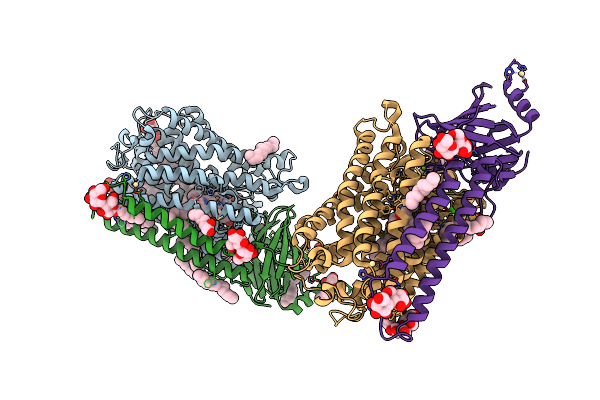 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: OH, HEA, DMU, TRD, HTH, CU, MG, CA, CD, TRS |
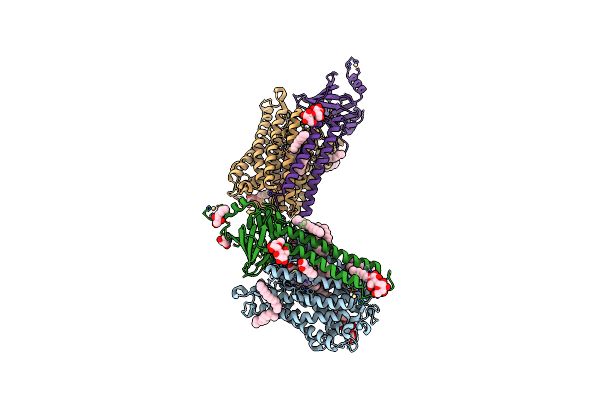 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: TRD, DMU, HEA, CU, MG, CA, HTH, TRS, CD |
 |
Catalytic Core Subunits (I And Ii) Of Cytochrome C Oxidase From Rhodobacter Sphaeroides With E101A (Ii) Mutation
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: DMU, TRD, CU, MG, CA, HEA, CD, HTH, TRS, K |
 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2017-09-13 Classification: OXIDOREDUCTASE Ligands: HEA, CU1, MG, CA, 3PE, LMU |
 |
R. Sphaeroides Photosythetic Reaction Center Mutant - Residue L223, Ser To Trp - Room Temperature Structure Solved On X-Ray Transparent Microfluidic Chip
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2017-04-12 Classification: PHOTOSYNTHESIS Ligands: BPH, BCL, FE, U10 |
 |
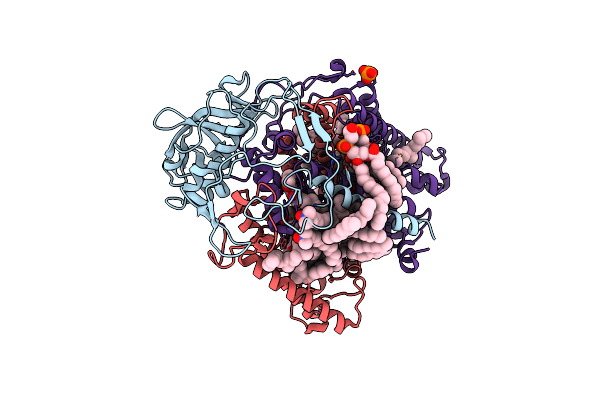
Photosynthetic Reaction Center Mutant With Glul212 Replaced With Trp (Chain L, El212W)
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-09 Classification: ELECTRON TRANSPORT Ligands: BCL, BPH, U10, DD9, FE, LDA, SPN, CDL |
 |
Photosynthetic Reaction Center Mutant With Glu L212 Replaced With Ala (Chain L, El212W), Asp L213 Replaced With Ala (Chain L, Dl213A) And Leu M215 Replaced With Ala (Chain M, Lm215A)
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-11-09 Classification: ELECTRON TRANSPORT Ligands: BCL, BPH, U10, D12, FE, SPN, LDA, PO4, CDL |
 |
Organism: Paracoccus denitrificans (strain pd 1222), Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-06-17 Classification: OXIDOREDUCTASE Ligands: CA, HEC |
 |
L(M196)H,H(M202)L Double Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-07-16 Classification: electron transport, membrane protein Ligands: LDA, UNL, PO4, DIO, EDO, TRS, K, BCL, BPH, U10, CDL, FE, SPN |
 |
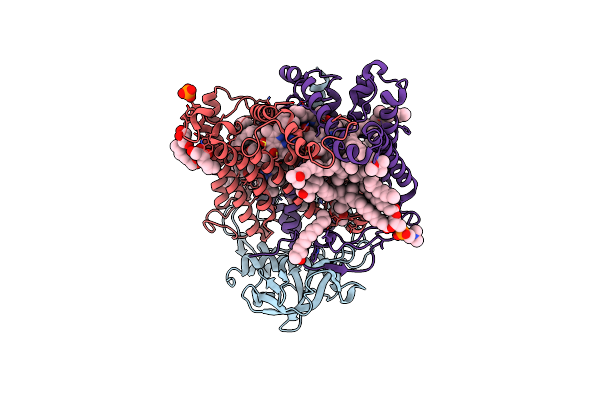
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE Ligands: CA, HEC, EDO, PO4, PGE, MES, NA |
 |
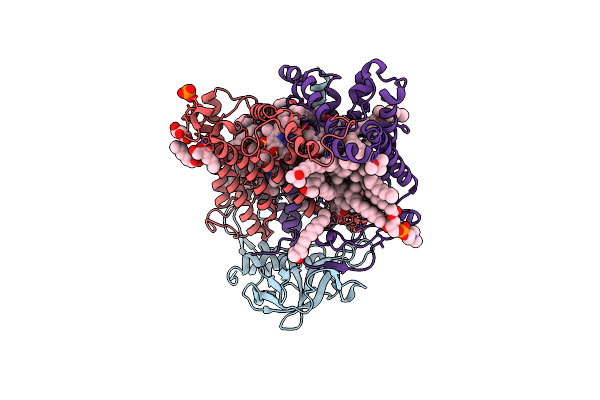
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2014-02-05 Classification: PHOTOSYNTHESIS Ligands: GOL, GGD, 2GO, LDA, U10, PO4, HTO, FE, SPO, CDL, PC1 |
 |
Zinc Substituted Reaction Center M(L214H) Variant Of Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2014-01-22 Classification: PHOTOSYNTHESIS Ligands: GOL, GGD, LDA, U10, 2GO, PO4, HTO, FE, SPO, CDL, PC1 |