Search Count: 15
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
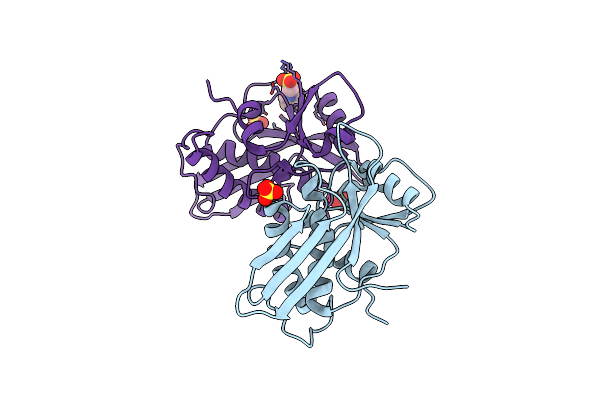
Crystal Structure Of The Apo Form Of D-Alanine: D-Alanine Ligase (Ddl) From Thermus Caldophilus: A Basis For The Substrate-Induced Conformational Changes
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-08-29 Classification: LIGASE |
 |
Crystal Structure And Functional Studies Reveal That Pas Factor From Vibrio Vulnificus Is A Novel Member Of The Saposin-Fold Family
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-02-14 Classification: LIPID BINDING PROTEIN |
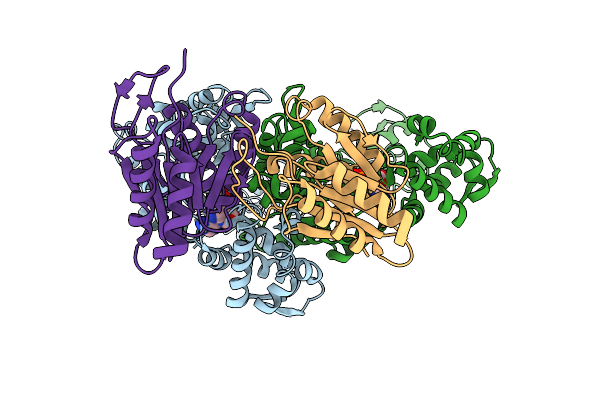 |
Correction Of X-Ray Intensities From An Hslv-Hslu Co-Crystal Containing Lattice Translocation Defects
Organism: Escherichia coli, Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:4.16 Å Release Date: 2005-07-12 Classification: CHAPERONE/HYDROLASE Ligands: ADP |
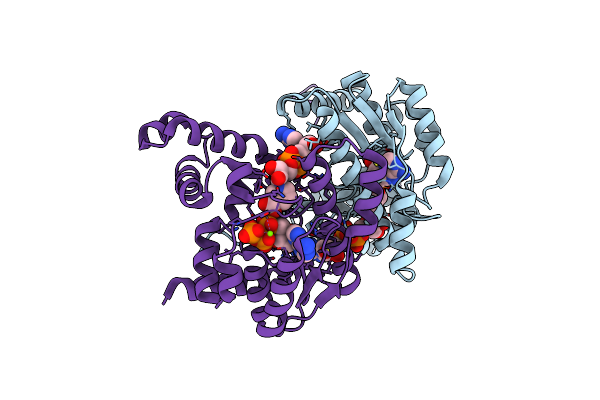 |
Crystal Structure Of Nh3-Dependent Nad+ Synthetase From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-04-05 Classification: LIGASE Ligands: MG, DND, ATP |
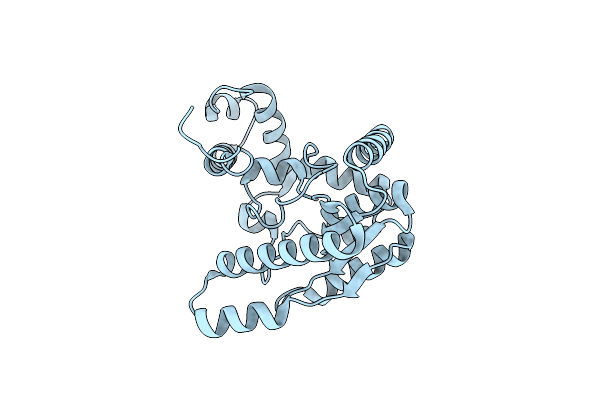 |
Crystal Structure Of Nh3-Dependent Nad+ Synthetase From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-04-05 Classification: LIGASE |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: SO4, MES |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-08-12 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The Sixth Pdz Domain Of Grip1 In Complex With Liprin C-Terminal Peptide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-08-12 Classification: PROTEIN BINDING |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-03-13 Classification: SIGNALING PROTEIN Ligands: SO4, MN |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-03-06 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2001-02-21 Classification: CHAPERONE/HYDROLASE Ligands: DAT |
 |
Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:7.00 Å Release Date: 2001-02-21 Classification: CHAPERONE/HYDROLASE |

