Search Count: 1,308
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: CA |
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: ADP |
 |
Proline Utilization A With The Fadh- N5 Atom Covalently Modified By Proline
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-06-04 Classification: OXIDOREDUCTASE Ligands: NAD, PGE, FDA, A1CAB, PRO, MG, SO4, A1AT4 |
 |
Proline Utilization A With The Covalent Acyl-Enzyme Intermediate In The Aldehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-06-04 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, PGE, A1AUG, MG, FDA, PEG |
 |
Proline Utilization A Complexed With The Substrate L-Glutamate Gamma-Semialdehyde In The Aldehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-06-04 Classification: OXIDOREDUCTASE Ligands: NAD, FDA, PGE, PEG, A1AUG, FMT, MG, SO4 |
 |
Proline Utilization A Complexed With The Product L-Glutamate In The Aldehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: GGL, SO4, PGE, FAD, PEG |
 |
Crystal Structure Of Sm0281, An Alpha/Beta Hydrolase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: GOL, PE3, CL |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Covalently Inactivated By (S)-But-3-Yn-2-Ylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: A1BC6, FMT |
 |
Proline Utilization A (Puta) From Sinorhizobium Meliloti Inactivated By N-Propargylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: P5F, NAD, SO4, MG, FMT, PEG, PG4, PGE |
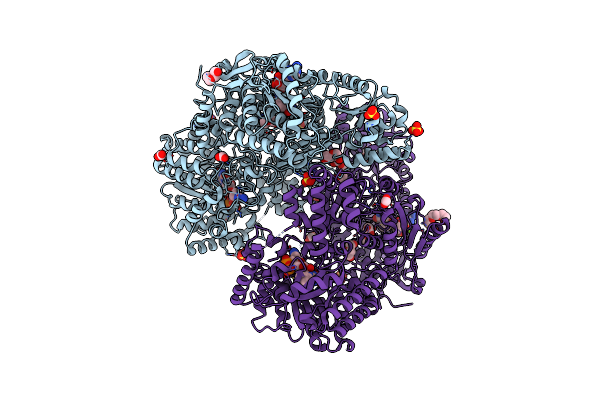 |
Structure Of Proline Utilization A Complexed With 2,3-Dihydro-1,4-Benzodioxine-5-Carboxylic Acid
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, 53E, FMT, NAD, SO4, MG, PGE, PG4 |
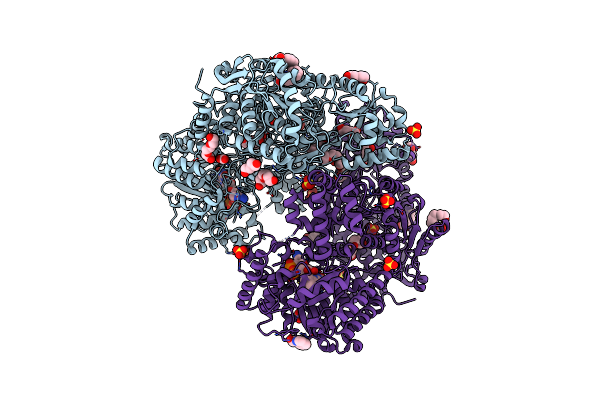 |
Structure Of Proline Utilization A Complexed With Quinoline-2-Carboxylic Acid
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, QNC, PEG, FMT, SO4, MG, PG4 |
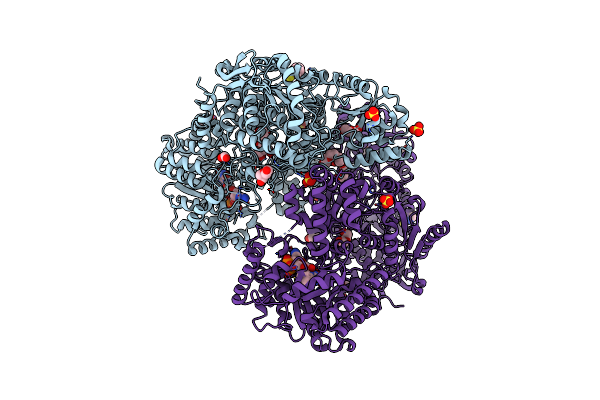 |
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: PGE, PEG, PYS, FMT, SO4, FAD |
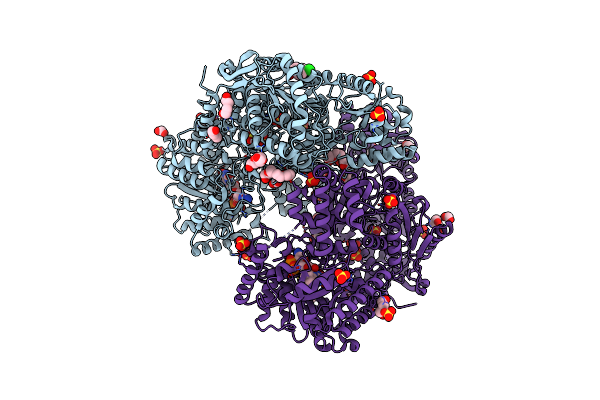 |
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, FMT, PEG, PG4, A1BDA, SO4 |
 |
Structure Of Proline Utilization A Complexed With Piperidine-3-Carboxylic Acid
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, PGE, FMT, A1BDB, ID7, SO4, NAD, MG, PG4 |
 |
Structure Of Proline Utilization A Complexed With 1-(4-Fluorophenyl)Thiourea
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, PGE, 8P4, FMT, SO4 |
 |
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, A1BDD, PGE, PG4, FMT, NAD, SO4, MG |
 |
Structure Of Proline Utilization A Co-Crystallized With 4-Methoxybenzyl Alcohol
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, A1BDD, PGE, FMT, PEG, NAD, SO4, MG, 1PE |
 |
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, FMT, PEG, NAD, SO4, MG, PGE, A1BDW |
 |
Structure Of Proline Utilization A Complexed With (2,3-Dihydro-1-Benzofuran-5-Yl)Methanol
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, FMT, A1AQC, NAD, SO4, MG, 1PE, PGE |
 |
Structure Of Proline Utilization A Complexed With 1-Benzofuran-5-Ylmethanol
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, FMT, A1BDX, NAD, SO4, MG, PGE, 1PE |

