Search Count: 7,998
All
Selected
 |
Crystal Structure Of Nodd-Ebd (Effector Binding Domain) In Complex With Hesperetin From Rhizobium Leguminosarum Bv. Vicae 3841
Organism: Rhizobium leguminosarum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: GOL, 6JP |
 |
Crystal Structure Of Nodd-Ebd (Effector Binding Domain) From Rhizobium Leguminosarum Bv. Vicae 3841
Organism: Rhizobium leguminosarum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-12-24 Classification: TRANSCRIPTION |
 |
Organism: Mesorhizobium metallidurans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: P6G, PEG, EPE, NA, TRS |
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: CA |
 |
Organism: Sinorhizobium meliloti
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: ADP |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: IMD, CL |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GDP, GOL, SO4 |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GFB, GOL |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: 5GP, DMS, FUC, A1CCV, BGC |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: 5GP, GOL, DMS, BGC, A1CCW, FUC, OC9 |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: 5GP, DMS, GMP, FUC, A1CCW |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: A1CCU, GLY, GOL, PO4 |
 |
Structure Of Phosphonate Monoester Hydrolase From Rhizobium Leguminosarum With Vanadate
Organism: Rhizobium johnstonii 3841
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MN, VO4, NA |
 |
Structure Of Phosphonate Monoester Hydrolase From Rhizobium Leguminosarum With Phenyl Phosphonate
Organism: Rhizobium johnstonii 3841
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MN, SV7, ACT, NA |
 |
The Outward-Open Structure Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
The Occluded Structures Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
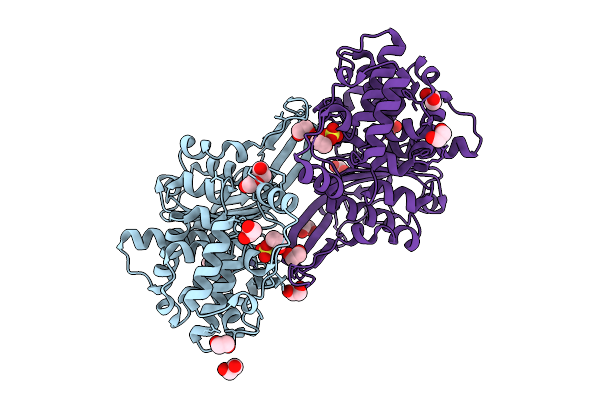
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-07-16 Classification: HYDROLASE Ligands: EDO, ZN, CL |
 |
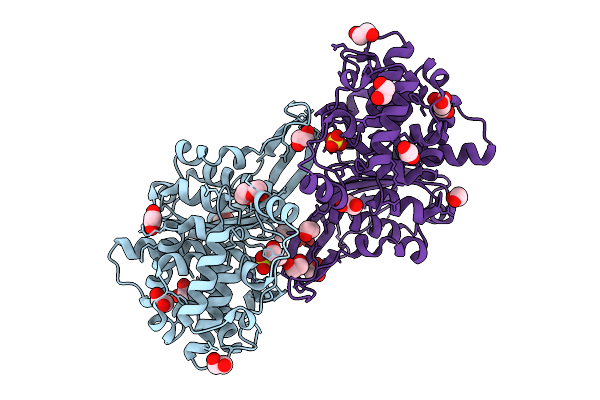
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-07-16 Classification: HYDROLASE Ligands: SO4, EDO, PEG, ZN, CL |
 |
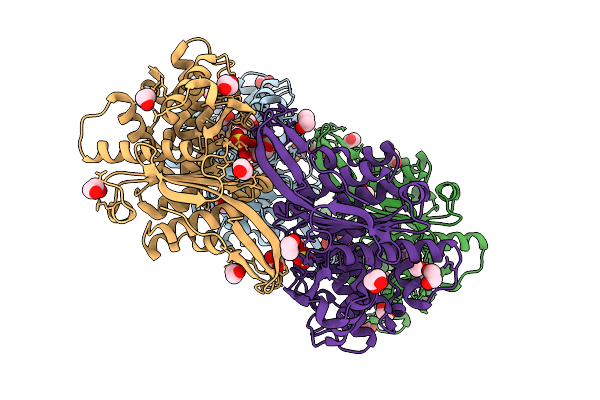
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-07-16 Classification: HYDROLASE Ligands: EDO, SO4, GOL, PEG, ZN |
 |
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE Ligands: SO4, PEG, EDO, ZN, CL |

