Search Count: 91
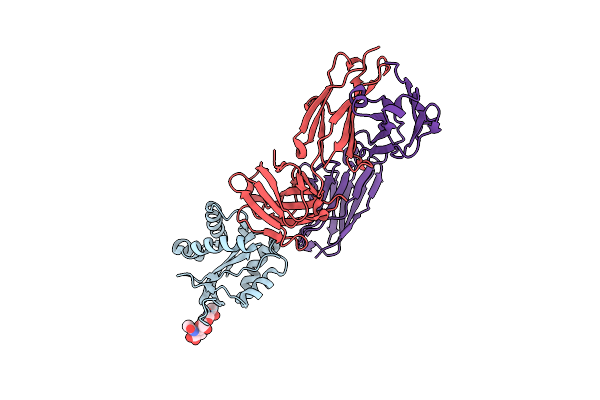 |
Crystal Structure Of Hiv-1 Mper Scaffold In Complex With Antibody Fab Ab45.1
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
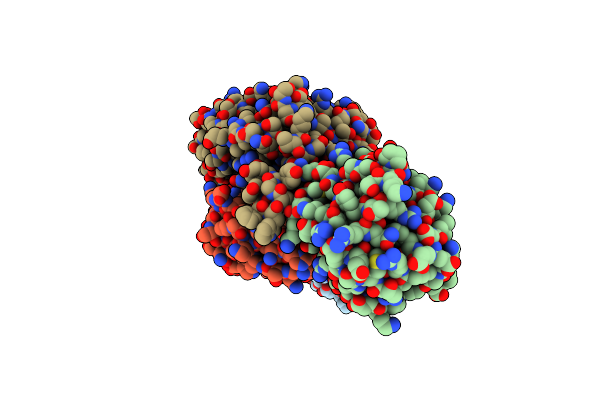 |
Crystal Structure Of Hiv-1 Mper Scaffold In Complex With Antibody Fab Ab45.2
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
 |
Organism: Plasmodium falciparum hb3, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-10-09 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Cryo-Em Structure Of Human Monoclonal Antibody C74 Targeting Pfd1235W (Cidra1.6) Pfemp1
Organism: Homo sapiens, Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: IMMUNE SYSTEM |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, CA, NA, PG4 |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: ASP, PG4, PGE, P6G, SAH, CA, NA |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, NA, CA, PG4, P6G |
 |
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: SAH, CA, PGE |
 |
Aeronamide N-Methyltransferase, Aere, Bound To Modified Peptide Substrate, Aera-Dl,34
Organism: Microvirgula aerodenitrificans dsm 15089
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-03-30 Classification: TRANSFERASE/BIOSYNTHETIC PROTEIN |
 |
Methanobactin Biosynthetic Protein Complex Of Mbnb And Mbnc From Methylosinus Trichosporium Ob3B At 2.62 Angstrom Resolution
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-03-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, SO4 |
 |
Methanobactin Biosynthetic Protein Complex Of Mbnb And Mbnc From Methylosinus Trichosporium Ob3B At 2.31 Angstrom Resolution
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-03-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, SO4 |
 |
Methanobactin Biosynthetic Protein Complex Of Mbnb And Mbnc From Methylosinus Trichosporium Ob3B, H210S Mutant
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-03-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, SO4 |
 |
Methanobactin Biosynthetic Protein Complex Of Mbnb And Mbnc From Methylosinus Trichosporium Ob3B At 2.21 Angstrom Resolution
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2022-03-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, SO4 |
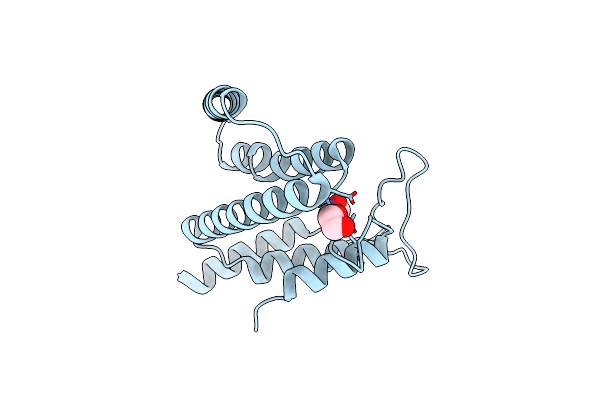 |
Crystal Structure Of A Putative Ntp Pyrophosphohydrolase (Exig_1061) From Exiguobacterium Sp. 255-15 At 1.78 A Resolution
Organism: Exiguobacterium sibiricum 255-15
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: EDO |
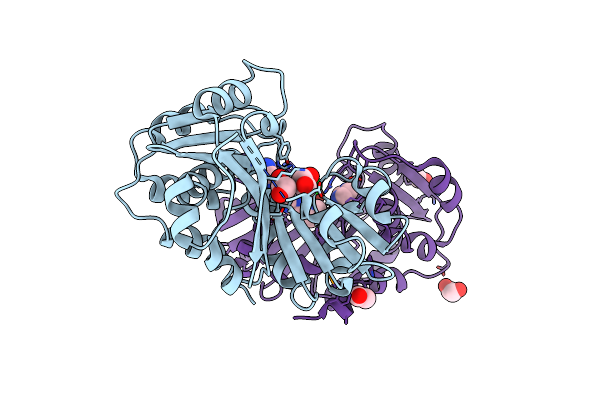 |
Crystal Structure Of Protein With Unknown Function From Duf364 Family (Zp_00559375.1) From Desulfitobacterium Hafniense Dcb-2 At 2.01 A Resolution
Organism: Desulfitobacterium hafniense
Method: X-RAY DIFFRACTION Release Date: 2010-02-02 Classification: Adenosyl binding protein Ligands: CL, IMD, EDO |
 |
Crystal Structure Of Putative Sufu (Suppressor Of Fused Protein) Homolog (Yp_208451.1) From Neisseria Gonorrhoeae Fa 1090 At 1.40 A Resolution
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-26 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Crystal Structure Of Putative Cell Invasion Protein With Mac/Perforin Domain (Np_812351.1) From Bacteriodes Thetaiotaomicron Vpi-5482 At 2.46 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2009-11-24 Classification: CELL INVASION Ligands: CL, EDO, MPD |
 |
Crystal Structure Of Protein With Unknown Function From Duf35 Family (13815350) From Sulfolobus Solfataricus At 1.80 A Resolution
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-09-01 Classification: acyl-CoA binding protein Ligands: ZN, SO4, ACY |
 |
Crystal Structure Of Pleckstrin Homology Domain (Yp_926556.1) From Shewanella Amazonensis Sb2B At 1.99 A Resolution
Organism: Shewanella amazonensis sb2b
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-06-23 Classification: structural genomics, unknown function Ligands: GOL, PEG |
 |
Crystal Structure Of A Putative Glycoside Hydrolase (Bt_2081) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.05 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-05-26 Classification: Ca-BINDING PROTEIN Ligands: CA, NA, CAC, ACT, PEG, P6G |

