Search Count: 24
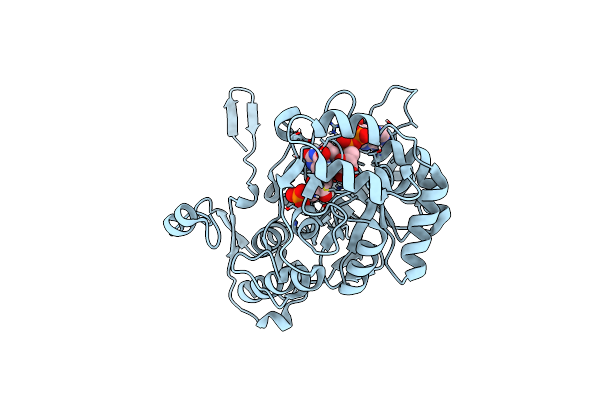 |
Structure A Catalytically Inactive Mutant Of The Imp Dehydrogenase Related Protein Guab3 From Synechocystis Pcc 6803
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-03-27 Classification: BIOSYNTHETIC PROTEIN Ligands: XMP, NAD |
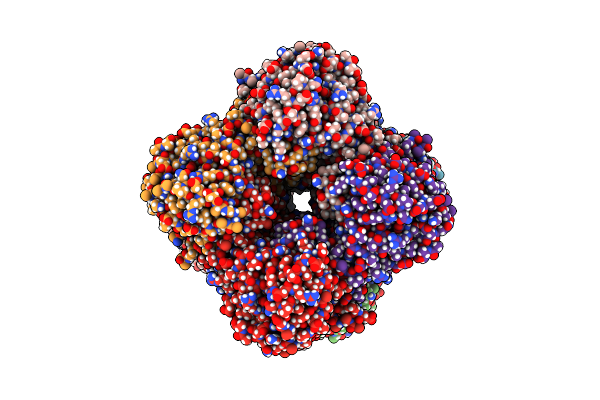 |
Structure Of The Imp Dehydrogenase Related Protein Guab3 From Synechocystis Pcc 6803
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-03-27 Classification: BIOSYNTHETIC PROTEIN Ligands: IMP, XMP |
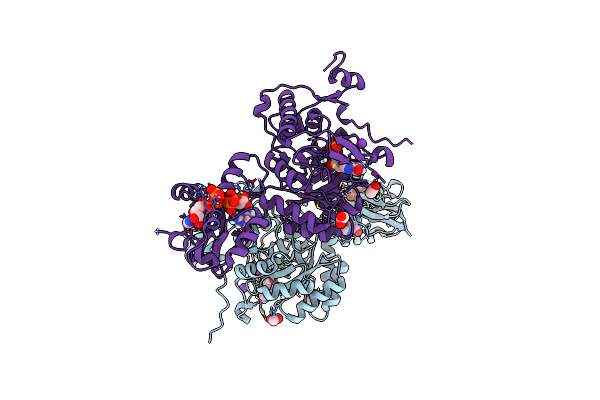 |
Crystal Structure Of Pseudomonas Aeruginosa Guab (Imp Dehydrogenase) Bound To Atp And Gdp At 1.65A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: ATP, GDP, IMP, MG, K, ACT |
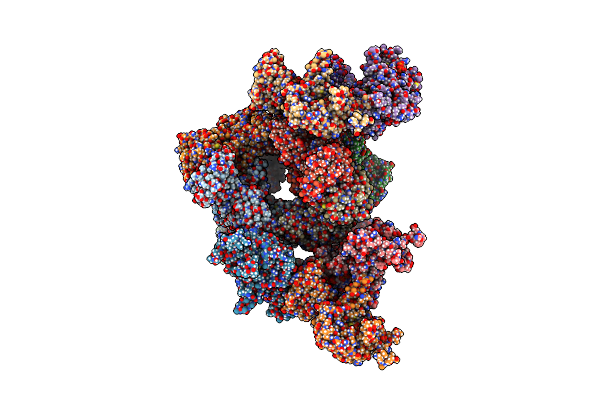 |
Crystal Structure Of Streptomyces Coelicolor Guab (Imp Dehydrogenase) Bound To Atp And Ppgpp At 2.0 A Resolution
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: ATP, G4P, MG |
 |
Structure Of The Ternary Complex Of The Impdh Enzyme From Ashbya Gossypii Bound To The Dinucleoside Polyphosphate Ap5G And Gdp
Organism: Eremothecium gossypii atcc 10895
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-08-28 Classification: OXIDOREDUCTASE Ligands: G5P, GDP, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2019-01-30 Classification: BIOSYNTHETIC PROTEIN Ligands: 5GP, GDP, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2019-01-30 Classification: BIOSYNTHETIC PROTEIN Ligands: GTP, SO4 |
 |
Crystal Structure Of A Thioredoxin From Clostridium Acetobutylicum At 1.75 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.3 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, PEG, ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.9 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, PEG, ACT |
 |
Crystal Structure Of A Ferredoxin-Flavin Thioredoxin Reductase From Clostridium Acetobutylicum At 1.64 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, PEG |
 |
Crystal Structure Of The Complex Of A Ferredoxin-Flavin Thioredoxin Reductase And A Thioredoxin From Clostridium Acetobutylicum At 2.9 A Resolution
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: FAD, EDO |
 |
Structure Of An Oxidoreductase Semet-Labelled From Synechocystis Sp. Pcc6803
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-11-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, EDO |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-11-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PG4, 1PE, PO4, PGE |
 |
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Release Date: 2017-11-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Release Date: 2017-11-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Organism: Gloeobacter violaceus (strain pcc 7421)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-07-27 Classification: OXIDOREDUCTASE Ligands: FAD, CA, PG4 |
 |
Organism: Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: 5GP, K, GOL |
 |
Organism: Ashbya gossypii (strain atcc 10895)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: 5GP, GDP, K, ACT |
 |
Structure Of The Covalent Intermediate E-Xmp* Of The Imp Dehydrogenase Of Ashbya Gossypii
Organism: Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: IMP |

