Search Count: 13
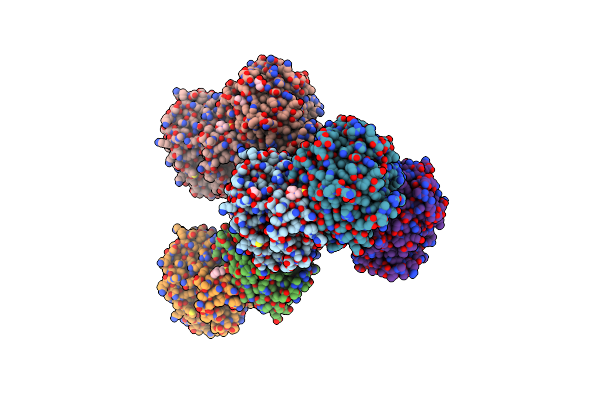 |
Crystal Structure Of Mgs-Mt1, An Alpha/Beta Hydrolase Enzyme From A Lake Matapan Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CL, MES, GOL |
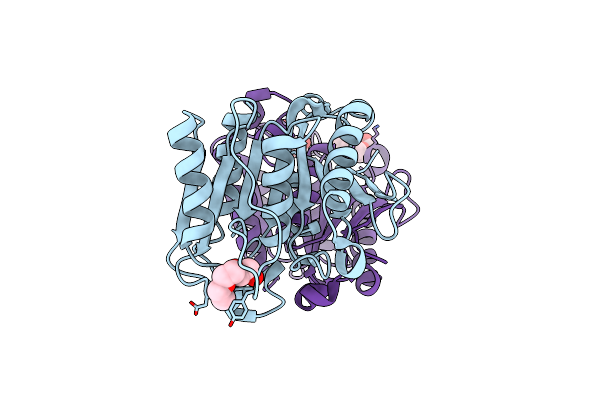 |
Crystal Structure Of Mgs-M1, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: CL, F, PGE |
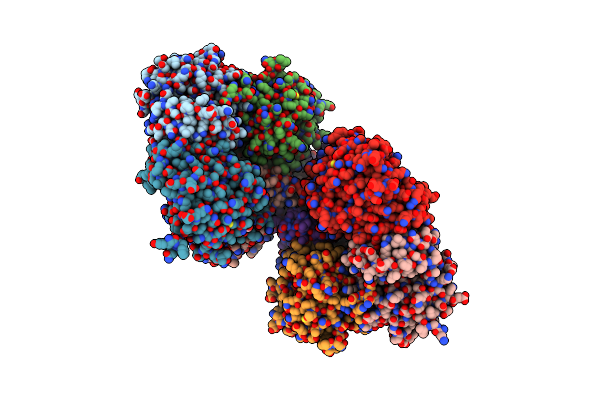 |
Crystal Structure Of Mgs-M2, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: GOL |
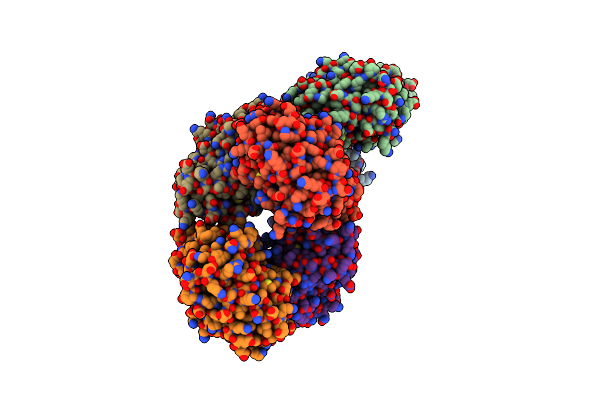 |
Crystal Structure Of Mgs-M4, An Aldo-Keto Reductase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: SO4, NA |
 |
Crystal Structure Of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-02-25 Classification: HYDROLASE |
 |
Crystal Structure Of A Putative Fumarylacetoacetate Isomerase/Hydrolase From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: HYDROLASE Ligands: ZN, TAR, ACT |
 |
Crystal Structure Of A Putative Kdpg (2-Keto-3-Deoxy-6-Phosphogluconate) Aldolase From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2012-01-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: PYR |
 |
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-13 Classification: HYDROLASE Ligands: SO4, NA, CL, CA |
 |
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2010-06-23 Classification: TRANSFERASE |
 |
The Crystal Structure Of The Protein Olei01261 With Unknown Function From Chlorobaculum Tepidum Tls
Organism: Oleispira antarctica rb-8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-03-16 Classification: Structural Genomics, Unknown function |
 |
Crystal Structure Of Amidohydrolase Family Protein Olei01672_1_465 From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-02-16 Classification: HYDROLASE Ligands: CA, ACY, PEG |
 |
Crystal Structure Of Phoshonoacetaldehyde Hydrolase Like Protein From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-09-01 Classification: structural genomics, unknown function |
 |
Structure Of A Putative Inorganic Pyrophosphatase From The Oil-Degrading Bacterium Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2009-07-28 Classification: HYDROLASE |

