Search Count: 13
 |
Organism: Salmonella enterica subsp. enterica serovar
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-08-26 Classification: Hydrolase, Transport protein Ligands: PG4, ETA, PEG, 1PE, MG, PGE, CA, P6G |
 |
Structure Of The F349A Mutant Of The Periplasmic Domain Of Yejm From Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-08-26 Classification: Hydrolase, Transport protein Ligands: P6G, PGE, PEG, ETA, EDO, PG4, MG, K, 1PE |
 |
Structure Of The Periplasmic Domain Of Yejm From Salmonella Typhimurium (Twinned)
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-08-26 Classification: Hydrolase, Transport protein Ligands: PGE, PEG, PO4, MN, EDO, PSE |
 |
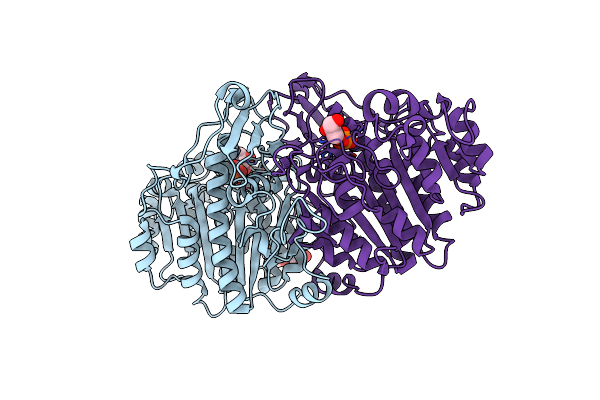
Crystal Structure Of Wild Type Alkaline Phosphatase Pafa To 1.7A Resolution
Organism: Elizabethkingia meningoseptica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of E. Coli Alkaline Phosphatase D101A/D153A In Complex With Inorganic Phosphate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2015-04-29 Classification: HYDROLASE Ligands: ZN, PO4, GOL |
 |
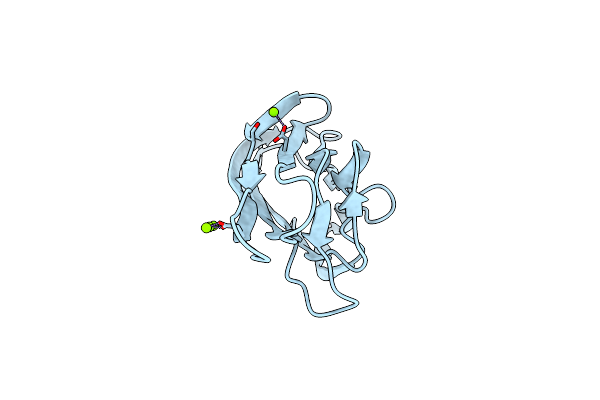
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2015-04-22 Classification: PROTEIN BINDING Ligands: MG |
 |
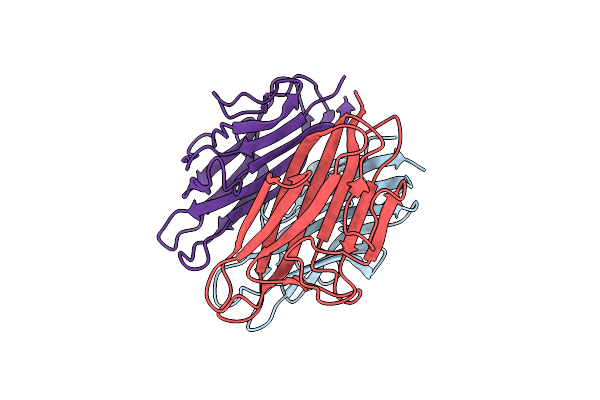
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-04-22 Classification: PROTEIN BINDING Ligands: MG |
 |
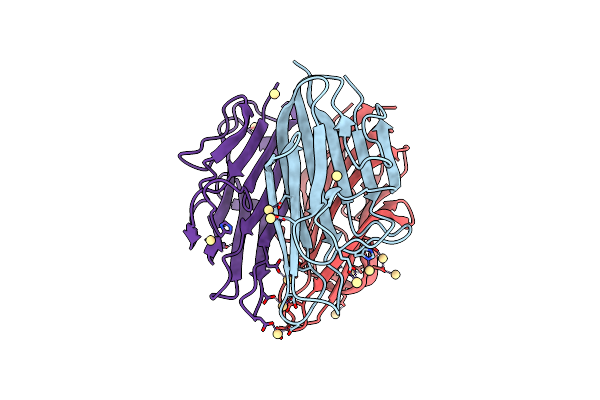
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2011-03-30 Classification: TRANSPORT PROTEIN Ligands: CHT |
 |
Crystal Structure Of The Sodium-Coupled Glycine Betaine Symporter Betp From Corynebacterium Glutamicum With Bound Substrate
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2009-05-26 Classification: MEMBRANE PROTEIN Ligands: BET |

