Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
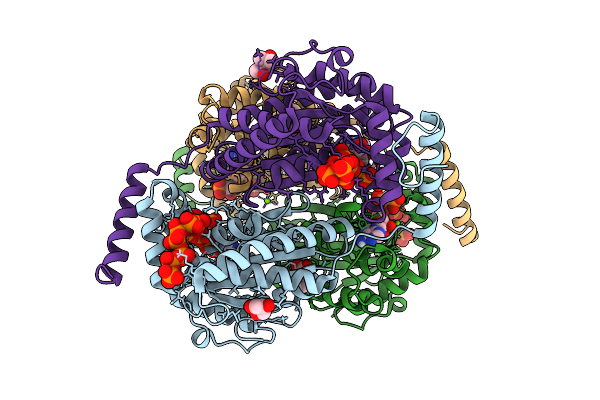 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Appch2P And Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: ACP, 6YY, GOL, BEZ, MG, MPD, A1I4D |
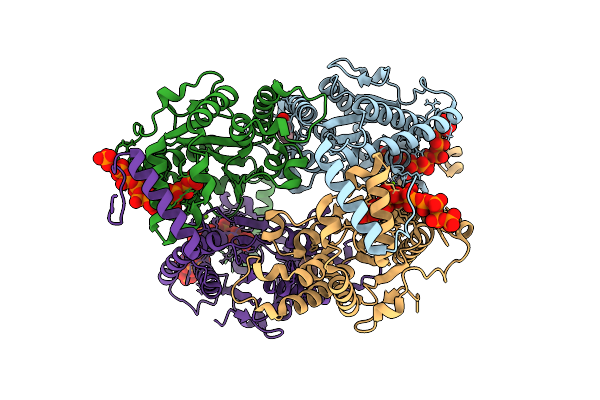 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: 6YY, BEZ, PO4, MG |
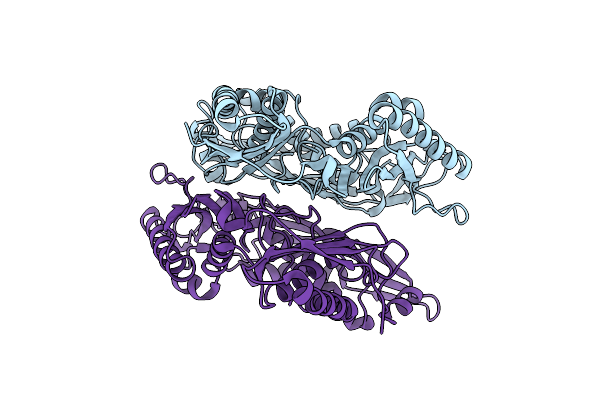 |
Crystal Structure Of Apo Form L147A/I351A Variant Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-11-17 Classification: TRANSFERASE |
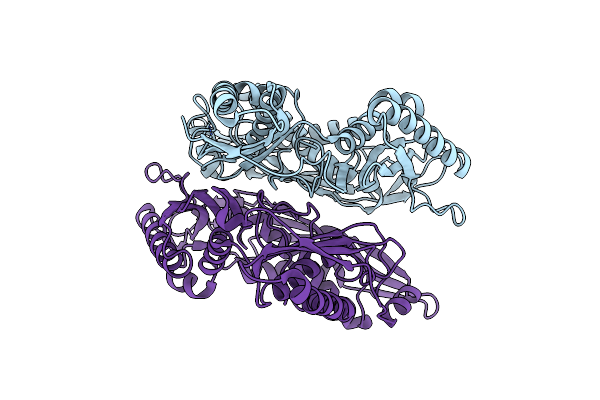 |
Crystal Structure Of Apo Form Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2021-11-17 Classification: TRANSFERASE |
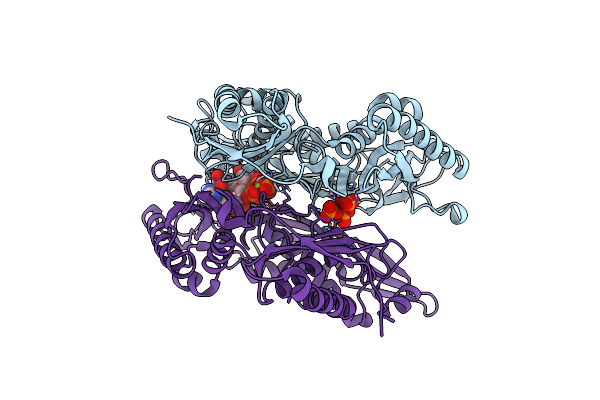 |
Crystal Structure Of L147A/I351A Variant Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii In Complex With Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-11-17 Classification: TRANSFERASE Ligands: 3PO, MG, EU9 |
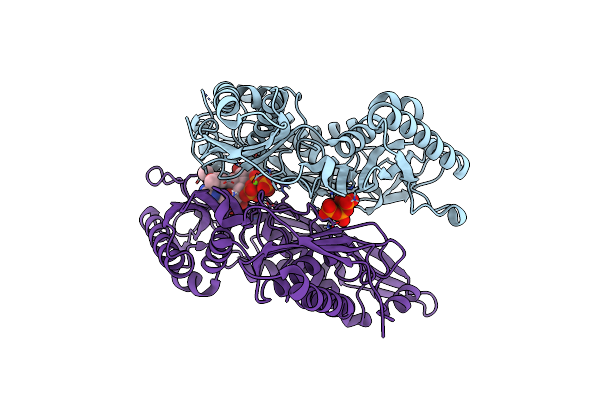 |
Crystal Structure Of L147A/I351A Variant Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii In Complex With Dmnb-Sam (4,5-Dimethoxy-2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-11-17 Classification: TRANSFERASE Ligands: 3PO, MG, 6IH |
 |
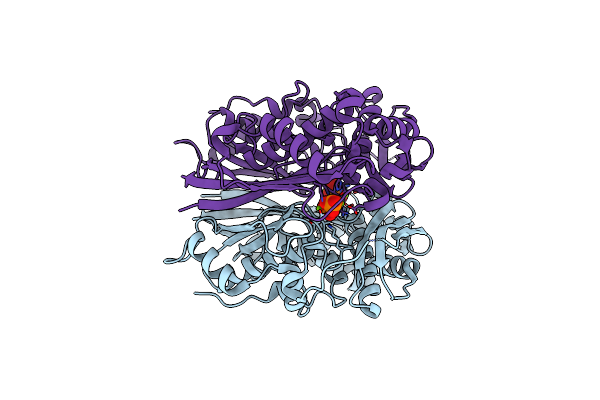
Crystal Structure Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis In Complex With Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: 3PO, MG, EU9 |
 |
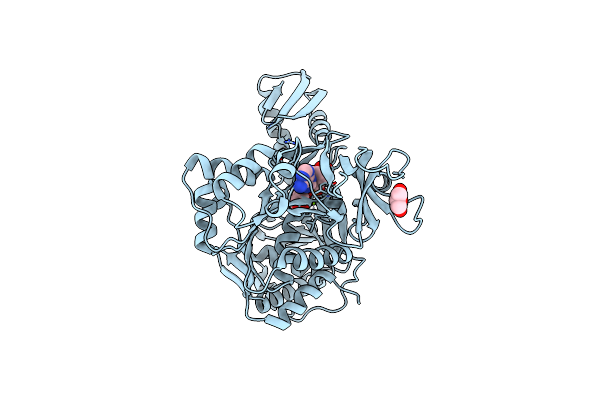
Crystal Structure Of Apo Form Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: MG, PO4 |
 |
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: ANP, GOL, MG |
 |
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: ARG, ANP, GOL, MG |
 |
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: TYR, PO4, GOL, AMP |
 |
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-01 Classification: TRANSCRIPTION Ligands: ARG, GOL, PO4, AMP |
 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: HEM |