Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
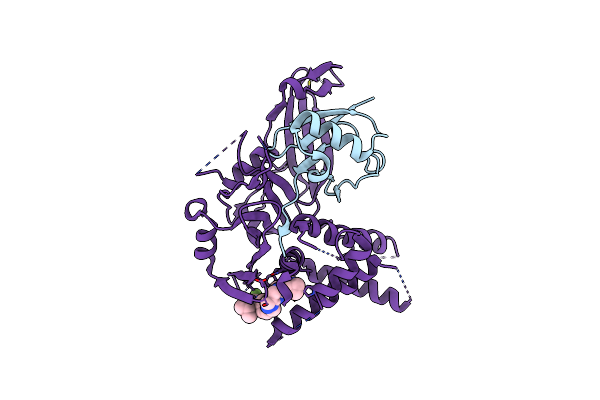 |
Usp1 Bound To Ksq-4279 And Ubiquitin Conjugated To Fancd2 (Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: HYDROLASE Ligands: ZN, A1IB8 |
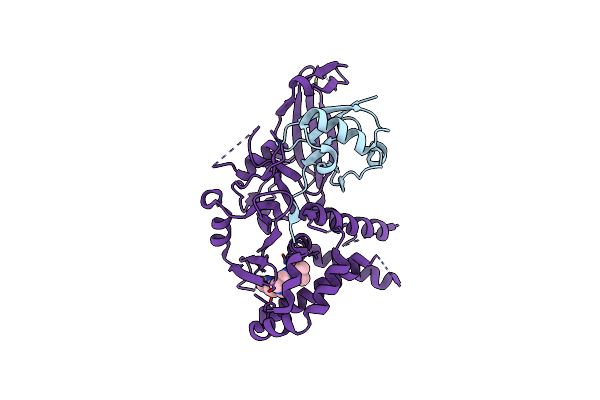 |
Usp1 Bound To Ml323 And Ubiquitin Conjugated To Fancd2 (Ordered Subset, Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: HYDROLASE Ligands: ZN, JDA |
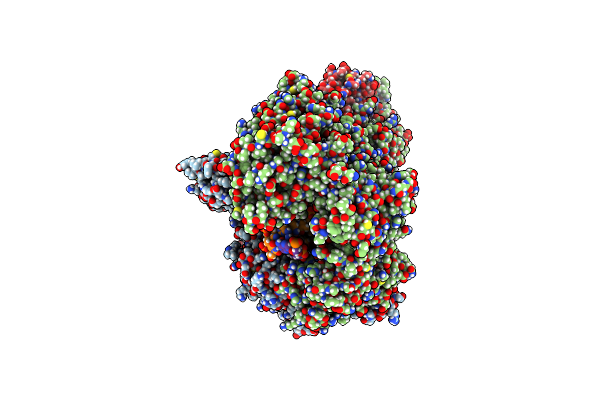 |
Structure Of Ubiquitinated Fanci In Complex With Fancd2 And Double-Stranded Dna
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN |
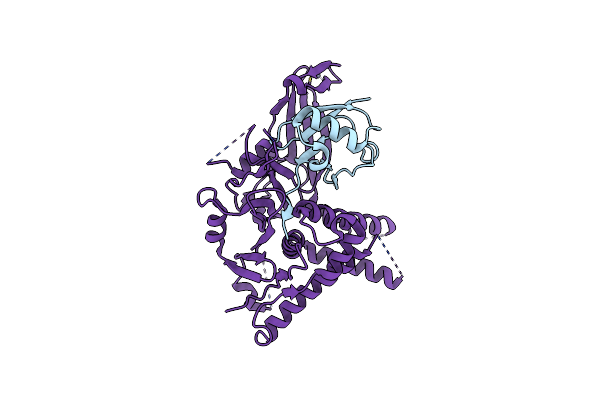 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZN |
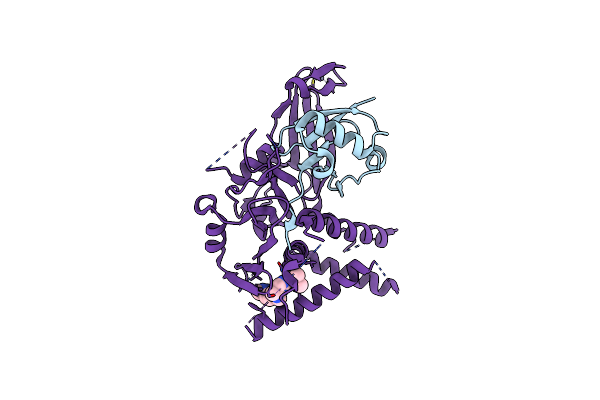 |
Usp1 Bound To Ml323 And Ubiquitin Conjugated To Fancd2 (Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZN, JDA |
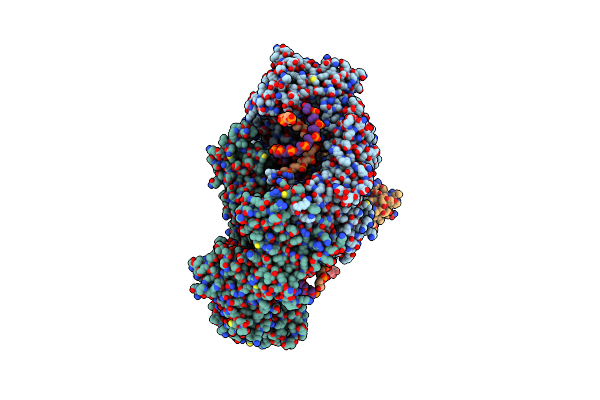 |
Cryo-Em Structure Of Usp1-Uaf1 Bound To Fanci And Mono-Ubiquitinated Fancd2 Without Ml323 (Consensus Reconstruction)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZN |
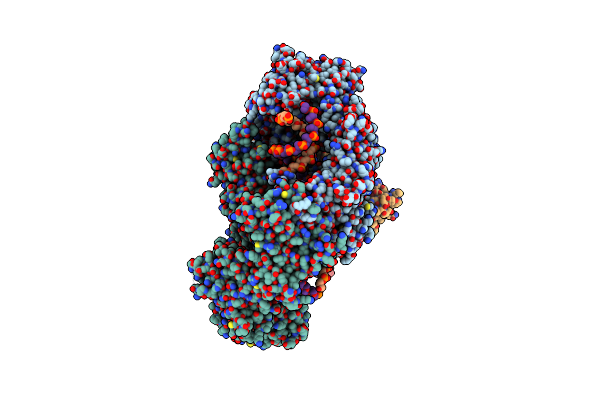 |
Cryo-Em Structure Of Usp1-Uaf1 Bound To Fanci And Mono-Ubiquitinated Fancd2 With Ml323 (Consensus Reconstruction)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZN, JDA |
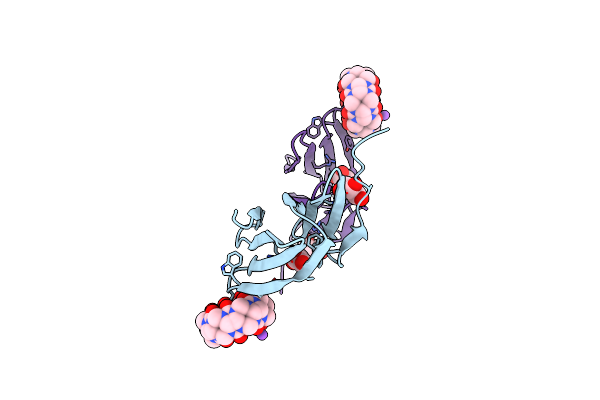 |
Dimethylated Fusion Protein Of Rsl And Mussel Adhesion Peptide (Mefp) In Complex With Cucurbit[7]Uril, H3 Sheet Assembly
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-09-08 Classification: SUGAR BINDING PROTEIN Ligands: QQ7, GOL, NA |
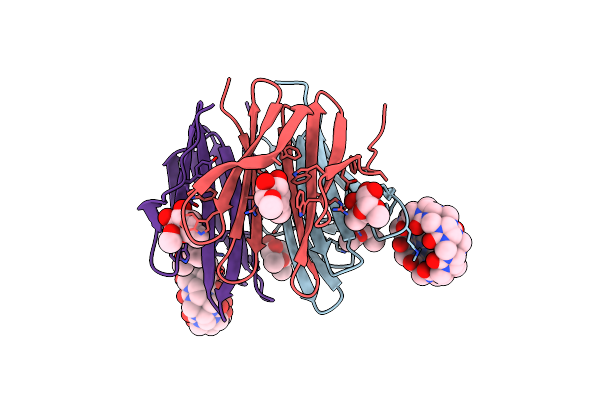 |
Dimethylated Fusion Protein Of Rsl And Nucleoporin Peptide (Nup) In Complex With Cucurbit[7]Uril, F432 Cage Assembly
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2021-09-08 Classification: SUGAR BINDING PROTEIN Ligands: MFU, QQ7 |
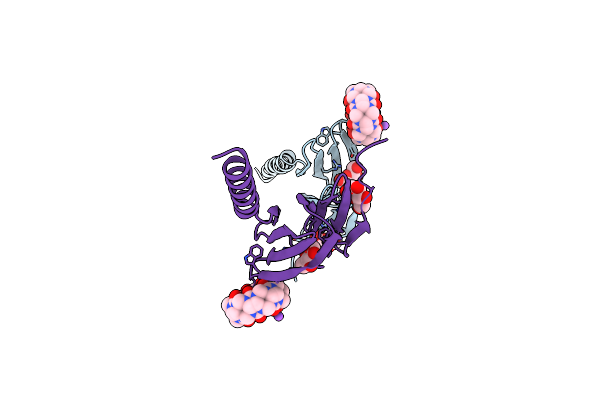 |
Dimethylated Fusion Protein Of Rsl And Trimeric Coiled Coil (4Dzn) In Complex With Cucurbit[7]Uril, H3 Sheet Assembly
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-09-08 Classification: SUGAR BINDING PROTEIN Ligands: QQ7, BMA, NA |
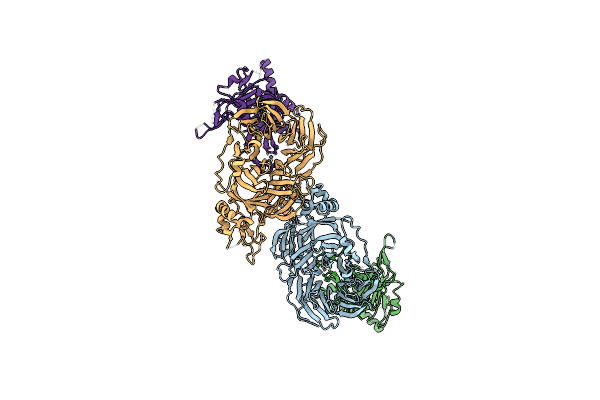 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2021-03-24 Classification: HYDROLASE Ligands: ZN |
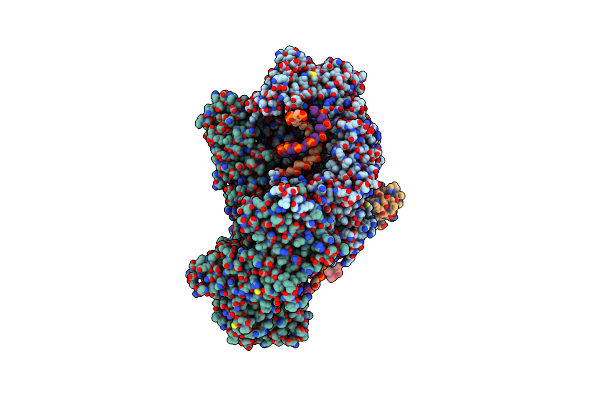 |
Cryo-Em Structure Of Usp1-Uaf1 Bound To Mono-Ubiquitinated Fancd2, And Fanci
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: HYDROLASE Ligands: ZN |
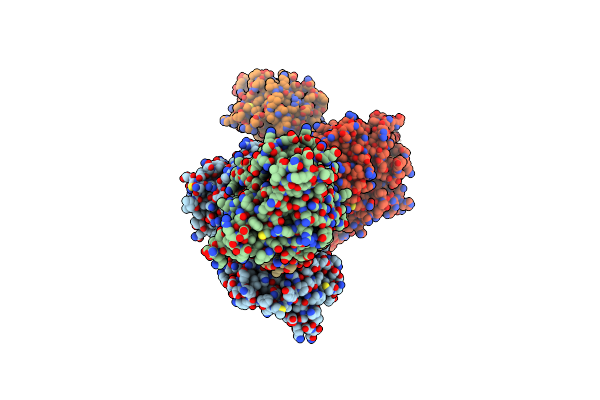 |
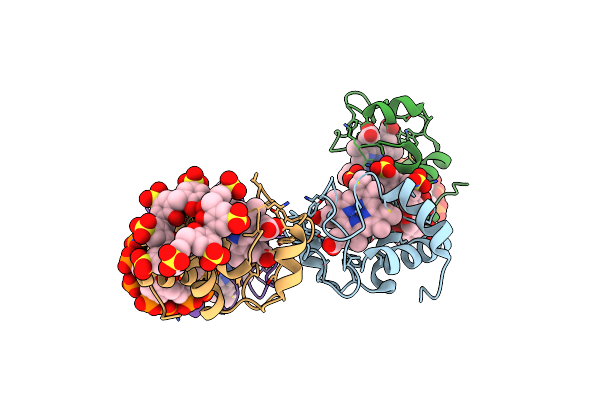
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-03-24 Classification: HYDROLASE Ligands: ZN, AYE |
 |
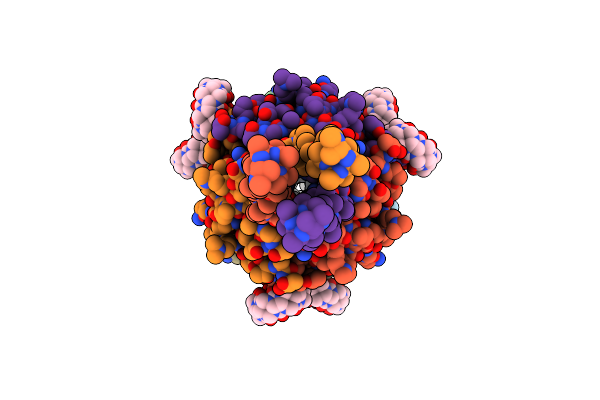
Cytochrome C In Complex With Phosphonato-Calix[6]Arene And Sulfonato-Calix[8]Arene
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEC, 7AZ, EVB |
 |
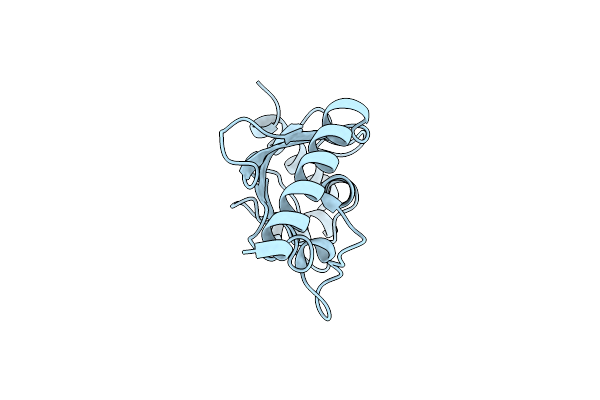
Dimethylated Fusion Protein Of Rsl And Trimeric Coiled Coil In Complex With Cucurbit[7]Uril
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-08-26 Classification: SUGAR BINDING PROTEIN Ligands: BMA, QQ7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-16 Classification: LIGASE |
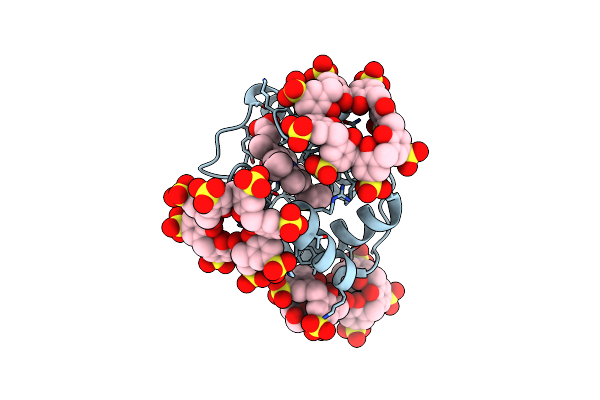 |
Cytochrome C Co-Crystallized With 25 Eq. Sulfonato-Calix[8]Arene - Structure 0
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: HEC, EVB, GOL |
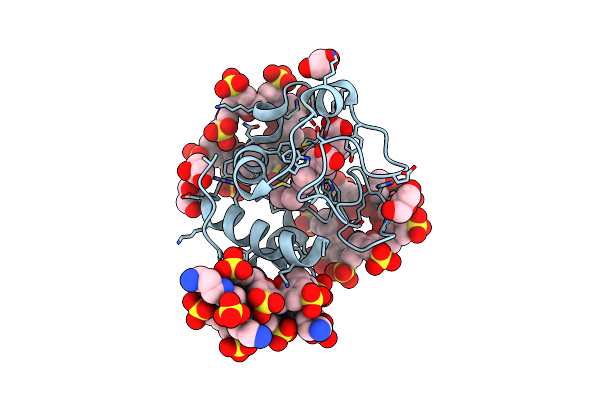 |
Cytochrome C Co-Crystallized With 20 Eq. Sulfonato-Calix[8]Arene And Soaked With 25 Eq. Spermine - Structure I
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: HEC, EVB, SPM, SO4, GOL |
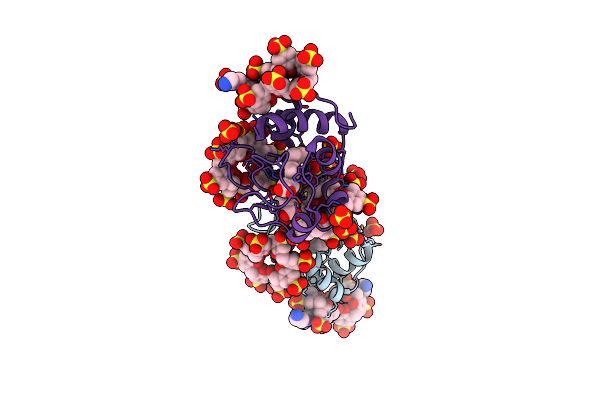 |
Cytochrome C Co-Crystallized With 20 Eq. Sulfonato-Calix[8]Arene And 15 Eq. Spermine - Structure Ii
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: HEC, EVB, SO4, SPM |
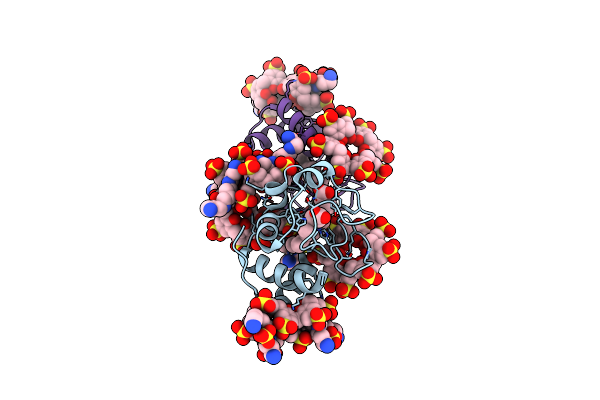 |
Cytochrome C Co-Crystallized With 10 Eq. Sulfonato-Calix[8]Arene And 25 Eq. Spermine (Dry-Coating Method) - Structure Iii
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: HEC, EVB, SPM, SO4 |