Search Count: 23
 |
Crystal Structure Of The Monomeric Avrm14-A Nudix Hydrolase Effector From Melampsora Lini
Organism: Melampsora lini
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-01-25 Classification: HYDROLASE |
 |
Crystal Structure Of The Monomeric Avrm14-B Nudix Hydrolase Effector From Melampsora Lini
Organism: Melampsora lini
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Homodimeric Avrm14-B Nudix Hydrolase Effector From Melampsora Lini
Organism: Melampsora lini
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-12-23 Classification: TRANSCRIPTION Ligands: QFN |
 |
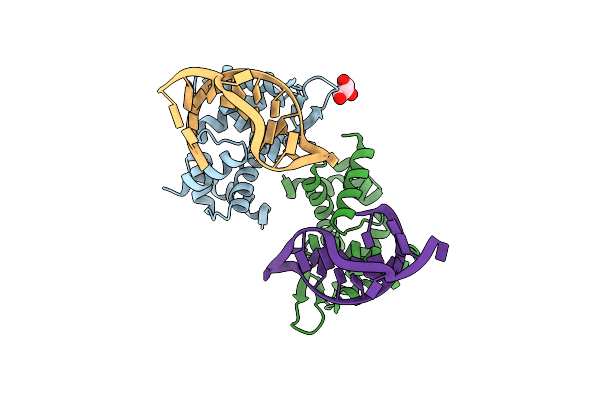
X-Ray Structure Of Roquin Roq Domain In Complex With A Selex-Derived Hexa-Loop Rna Motif
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-04-06 Classification: RNA BINDING PROTEIN |
 |
X-Ray Structure Of Roquin Roq Domain In Complex With Ox40 Hexa-Loop Rna Motif
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2016-04-06 Classification: RNA BINDING PROTEIN Ligands: GOL |
 |
The Structure Of A Yeast Dynein Dyn2-Pac11 Complex And Effect On Single Molecule Dynein Motor Activity
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-09-18 Classification: MOTOR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OK |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: NAR, IRG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0ON |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: IRG, 0OQ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-12-05 Classification: Transferase/Transferase inhibitor Ligands: 0SJ, IRG |
 |
Crystal Structure Of The Complex Formed Between Alkaline Proteinase Savinase And Gramicidin S At 1.5A Resolution
Organism: Brevibacillus brevis, Bacillus lentus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2004-06-22 Classification: HYDROLASE/ANTIBIOTIC Ligands: CA |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2004-05-11 Classification: HYDROLASE |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-05-11 Classification: HYDROLASE Ligands: SM |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-05-11 Classification: HYDROLASE Ligands: MG, ATP |

