Search Count: 59
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MG, PO4, UK9, PGE, CIT, GOL |
 |
Human Pyridoxal Phosphatase In Complex With 7,8-Dihydroxyflavone Without Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: UK9, FMT, MG, CL |
 |
Human Pyridoxal Phosphatase In Complex With 7,8-Dihydroxyflavone And Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MG, UK9, GOL, PO4 |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Uman metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Rigid Body Fit Of Assembled Hmpv N-Rna Spiral Bound To The C-Terminal Region Of P
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
Organism: Human metapneumovirus (strain can97-83), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRAL PROTEIN |
 |
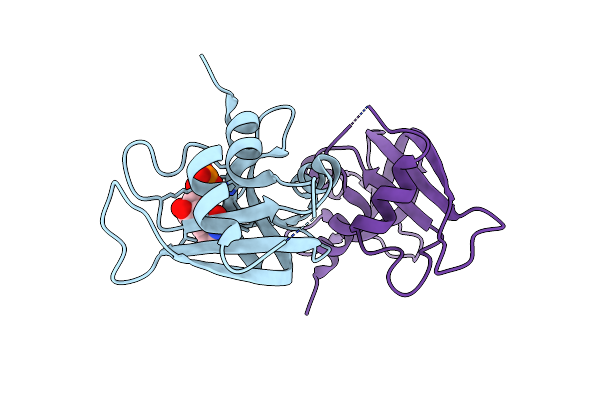
Structure Of A2A Adenosine Receptor A2Ar-Star2-Bril, Solved At Wavelength 2.75 A
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: TEP, OLA, CLR |
 |
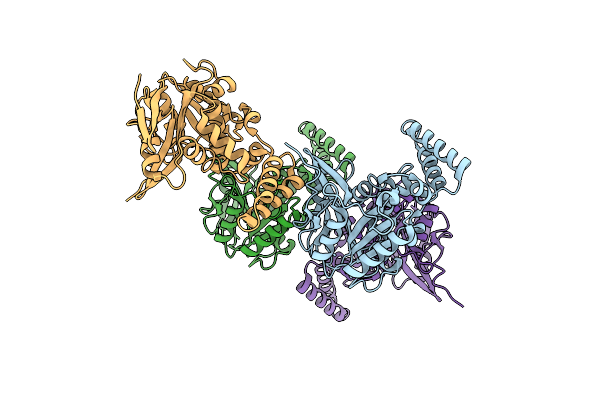
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: 0U |
 |
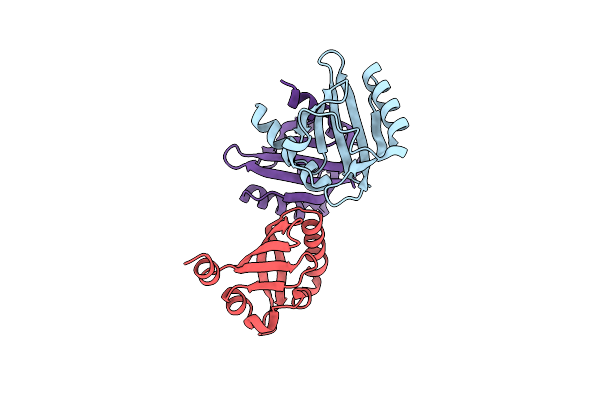
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-10-25 Classification: TOXIN |
 |
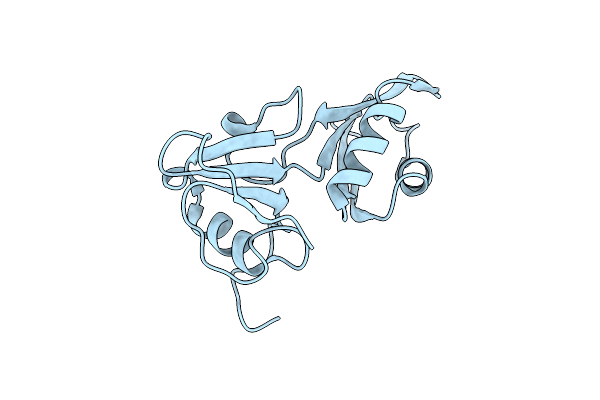
Structure Of The Pas Domain Code By The Lic_11128 Gene From Leptospira Interrogans Serovar Copenhageni Fiocruz, Solved At Wavelength 3.09 A
Organism: Leptospira interrogans serovar copenhageni
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION |
 |
Structure Of The Rna Recognition Motif (Rrm) Of Seb1 From S. Pombe., Solved At Wavelength 2.75 A
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-10-25 Classification: TRANSCRIPTION |
 |
Structure Of Bacterial Multidrug Efflux Transporter Acrb, Solved At Wavelength 3.02 A
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Antibacterial Peptide Abc Transporter Mcjd, Solved At Wavelength 2.75 A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: MG, ANP |
 |
Structure Of Fap1, A Domain Of The Accessory Sec-Dependent Serine-Rich Glycoprotein Adhesin From Streptococcus Oralis, Solved At Wavelength 3.06 A
Organism: Streptococcus oralis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-10-25 Classification: CELL ADHESION Ligands: CA, EDO |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-25 Classification: PEPTIDE BINDING PROTEIN Ligands: CL, GOL |

